LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis - PubMed (original) (raw)
LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis
Rita Gross-Hardt et al. PLoS Biol. 2007 Mar.
Abstract
In flowering plants, the egg and sperm cells form within haploid gametophytes. The female gametophyte of Arabidopsis consists of two gametic cells, the egg cell and the central cell, which are flanked by five accessory cells. Both gametic and accessory cells are vital for fertilization; however, the mechanisms that underlie the formation of accessory versus gametic cell fate are unknown. In a screen for regulators of egg cell fate, we isolated the lachesis (lis) mutant which forms supernumerary egg cells. In lis mutants, accessory cells differentiate gametic cell fate, indicating that LIS is involved in a mechanism that prevents accessory cells from adopting gametic cell fate. The temporal and spatial pattern of LIS expression suggests that this mechanism is generated in gametic cells. LIS is homologous to the yeast splicing factor PRP4, indicating that components of the splice apparatus participate in cell fate decisions.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
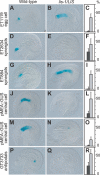
Figure 1. Accessory Cells in lis-1 Gametophytes Morphologically Resemble Gametic Cells
(A–D) Schematic representation of wild-type female gametophyte development. Sporophytic structures are shown in grey; gametophytic structures are colored. (A) After meiosis, the haploid functional megaspore is formed. (B) A series of three mitotic divisions results in the formation of an eight-nucleate syncytium. (C) After nuclear migration and cellularization, a seven-celled gametophyte is formed containing two synergids at the micropylar end (dark green), one egg cell (red), one central cell (orange) with two polar nuclei, and three antipodal cells at the chalazal pole (light green). (D) Prior to fertilization, the two polar nuclei fuse to form one large central cell nucleus, and the antipodal cells degenerate. (E) Wild-type silique showing full seed set. (F) Silique of lis-1/LIS plants containing aborted ovules (arrowheads). (G–I) Mature wild-type gametophyte. (G) At the micropylar end, the two small synergid nuclei are detected (stars). The larger egg cell nucleus (arrowhead) is oriented towards the adjacent central cell. (H) A large central cell nucleus (arrowhead) resulting from the fusion of the two polar nuclei can be detected. (I) The antipodal cells at the chalazal end degenerate (star). (J–N) lis-1 gametophytes. (J) The synergid nuclei are enlarged and mis-polarized (star). As a consequence, synergid and egg cell become indistinguishable when lying in a similar position (arrowheads). (K) Polar nuclei are unfused (arrowheads) and occasionally ectopically cellularized (arrowheads in M). (L and M) Antipodal cells do not degenerate, but enlarge and protrude towards the center (stars). (N) Disintegration of antipodal cells and fused antipodal nuclei (star).
Figure 2. Molecular Analysis of lis-1 and Wild-Type Gametophytes
Expression analysis of cell-specific marker genes in wild-type versus lis-1/LIS plants (2 d after emasculation if not otherwise indicated). The onset of GUS expression for all markers is only detected after cellularization. (A, D, G, J, M, and P) The wild-type expression pattern for each marker. (B, E, H, K, N, and Q) Abnormal expression patterns, representing the scored phenotypes. (C, F, I, L, O, and R) Frequencies of abnormal patterns. Dark bars represent wild-type, light bars represent lis-1/LIS plants. The _y_-axis shows the percentage of the scored phenotype. (A–C) The egg cell marker ET1119 was expressed ectopically in 36.4% of the gametophytes of lis-1/LIS plants (n = 562; wild-type: 1.2%, n = 423). (D–F) The synergid marker ET2634 was not expressed in 41.5% of lis-1/LIS gametophytes (n = 789; wild-type: 22.0%, n = 1,176). (G–I) The synergid marker ET884 was expressed ectopically in 38.7% of the gametophytes of lis-1/LIS plants (n = 694; wild-type: 0.1%, n = 1,055). (J–L) Expression of the central cell pMEA::GUS reporter construct was not detected in 47.8% of the gametophytes of lis-1/LIS plants (n = 596; wild-type: 10.7%, n = 428). (M–O) Ectopic expression of pMEA::GUS reporter construct in antipodal cells 3 d after emasculation was detected in 16.4% of the gametophytes of lis-1/LIS plants (n = 531; wild-type: 1.3%, n = 468). (P–R) The antipodal marker GT3733 was not expressed in 53.1% of the gametophytes of lis-1/LIS plants (n = 715; wild-type: 22.2%, n = 1,283).
Figure 3. Functional Analysis of Synergids and Central Cells in lis-1/LIS Plants
(A–C) GUS staining in synergids after fertilization of wild-type and lis-1/LIS plants with pollen from the ET434G pollen-tube marker line. (A) Ovule with GUS-stained synergids. The arrowhead points at pollen tube. (B) Ovule in which no GUS staining was detected in synergids. (C) Frequencies of GUS negative synergids. Dark bars represent wild-type, light bars represent lis-1/LIS plants. The _y_-axis shows the percentage of the scored phenotype (lis-1/LIS: 50.8%, n = 789; wild-type: 21.5%, n = 287). (D–F) Endosperm development after fertilization of wild-type and lis-1/LIS plants with wild-type pollen. (D) Ovule with developing embryo (star) and endosperm (arrowhead). (E) Ovule with a developing embryo (star), but no endosperm. The undeveloped central cell nucleus is visible (arrowhead). (F) Frequencies of ovules with a developing embryo, but no endosperm. Dark bars represent wild-type, light bars represent lis-1/LIS plants. The _y_-axis shows the percentage of the scored phenotype (lis-1/LIS: 11.2%, n = 267; wild type: 0.5%, n = 191).
Figure 4. Successive Recruitment of Cells as Gametic Cells in lis-1 Gametophytes
(A and B) Ectopic expression of the egg cell marker ET1119 (A) and the central cell marker pMEA::GUS (B) in wild-type and lis-1/LIS plants. Dark bars represent wild-type, light bars represent lis-1/LIS plants. The _y_-axis shows the percentage of ectopic expression of total GUS-staining ovules. Ectopic expression was scored zero (0d), one (1d), and two days (2d) after emasculation. Total number of ovules counted was greater than 250. (C–G) Development of five morphological features in lis-1/LIS plants as compared to wild type, zero (0d), one (1d), and two days (2d) after emasculation. The _y_-axis shows percent deviation from wild type (data from Table S1). (C) Synergid nuclei smaller than egg cell nucleus. (D) Different polarity of synergids and egg cell with respect to position of nucleus. (E) Polar nuclei unfused. (F) Ectopic cellularization. (G) Protruded antipodal cells.
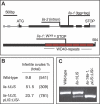
Figure 5. LIS Codes for a WD40 Repeat Protein
(A) Gene (upper panel) and protein (lower panel) structure of LIS. The localization of the lis-1 and lis-2 mutations and the seven WD40 repeats are indicated. (B and C) LIS cDNA driven by a 2.6-kb upstream promoter complements the gametophytic (B) as well as possible sporophytic defects of lis-1/lis-1 mutants, allowing for the generation of homozygous plants, as demonstrated by PCR-based genotyping (C).
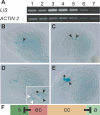
Figure 6. LIS Is Strongly Expressed in Gametic Cells
(A) RT-PCR analysis of LIS expression in leaves (1), roots (2), flower buds (3), open flowers (4), inflorescences (5), siliques (6), and stem (7) (upper panel). ACTIN 2 was used as control (lower panel). (B–E), Expression of pLIS::NLS_GUS in wild-type ovules during female gametophyte development. (B) Arrowhead points at the functional megaspore in which GUS expression is detected in the nucleus. (C) Two-nucleate embryo sac showing GUS expression in both nuclei (arrowheads). (D) Young eight-nucleate gametophyte. Antipodal nuclei are not visible. Inset shows area with GUS-positive nuclei at higher magnification. White arrowheads point to the synergid nuclei; star points to the egg cell nucleus; and black arrowheads point to the unfused polar nuclei. (E) Mature gametophyte showing strong GUS signal in both the egg cell (star) and fused central cell nucleus (arrowhead). Expression in synergid cells is hardly detectable. (F) Proposed model for LIS function. Expression of LIS in gametic cells (egg cell [ec] and central cell [cc]) is necessary for the generation of a lateral inhibition signal that prevents the adjacent accessory cells (synergids [s] and antipodal cells [a]) from adopting gametic cell fate.
Similar articles
- CLO/GFA1 and ATO are novel regulators of gametic cell fate in plants.
Moll C, von Lyncker L, Zimmermann S, Kägi C, Baumann N, Twell D, Grossniklaus U, Gross-Hardt R. Moll C, et al. Plant J. 2008 Dec;56(6):913-21. doi: 10.1111/j.1365-313X.2008.03650.x. Epub 2008 Sep 12. Plant J. 2008. PMID: 18702672 - LACHESIS-dependent egg-cell signaling regulates the development of female gametophytic cells.
Völz R, von Lyncker L, Baumann N, Dresselhaus T, Sprunck S, Gross-Hardt R. Völz R, et al. Development. 2012 Feb;139(3):498-502. doi: 10.1242/dev.075234. Epub 2011 Dec 21. Development. 2012. PMID: 22190635 - Mutants with aberrant numbers of gametic cells shed new light on old questions.
Moll C, Nielsen N, Gross-Hardt R. Moll C, et al. Plant Biol (Stuttg). 2008 Sep;10(5):529-33. doi: 10.1111/j.1438-8677.2008.00127.x. Plant Biol (Stuttg). 2008. PMID: 18761491 - How females become complex: cell differentiation in the gametophyte.
Kägi C, Gross-Hardt R. Kägi C, et al. Curr Opin Plant Biol. 2007 Dec;10(6):633-8. doi: 10.1016/j.pbi.2007.07.011. Epub 2007 Sep 11. Curr Opin Plant Biol. 2007. PMID: 17851110 Review. - Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis.
Liu J, Qu LJ. Liu J, et al. Mol Plant. 2008 Jul;1(4):564-74. doi: 10.1093/mp/ssn033. Mol Plant. 2008. PMID: 19825562 Review.
Cited by
- TMT-Based Quantitative Proteomic Analysis Reveals the Crucial Biological Pathways Involved in Self-Incompatibility Responses in Camellia oleifera.
He Y, Song Q, Wu Y, Ye S, Chen S, Chen H. He Y, et al. Int J Mol Sci. 2020 Mar 14;21(6):1987. doi: 10.3390/ijms21061987. Int J Mol Sci. 2020. PMID: 32183315 Free PMC article. - Two SUMO Proteases SUMO PROTEASE RELATED TO FERTILITY1 and 2 Are Required for Fertility in Arabidopsis.
Liu L, Jiang Y, Zhang X, Wang X, Wang Y, Han Y, Coupland G, Jin JB, Searle I, Fu YF, Chen F. Liu L, et al. Plant Physiol. 2017 Dec;175(4):1703-1719. doi: 10.1104/pp.17.00021. Epub 2017 Oct 24. Plant Physiol. 2017. PMID: 29066667 Free PMC article. - Rice MutLγ, the MLH1-MLH3 heterodimer, participates in the formation of type I crossovers and regulation of embryo sac fertility.
Mao B, Zheng W, Huang Z, Peng Y, Shao Y, Liu C, Tang L, Hu Y, Li Y, Hu L, Zhang D, Yuan Z, Luo W, Yuan L, Liu Y, Zhao B. Mao B, et al. Plant Biotechnol J. 2021 Jul;19(7):1443-1455. doi: 10.1111/pbi.13563. Epub 2021 Feb 22. Plant Biotechnol J. 2021. PMID: 33544956 Free PMC article. - Morphological and Physiological Framework Underlying Plant Longevity in Arabidopsis thaliana.
Wang Y, Kumaishi K, Suzuki T, Ichihashi Y, Yamaguchi N, Shirakawa M, Ito T. Wang Y, et al. Front Plant Sci. 2020 Nov 5;11:600726. doi: 10.3389/fpls.2020.600726. eCollection 2020. Front Plant Sci. 2020. PMID: 33224176 Free PMC article.
References
- Dresselhaus T. Cell-cell communication during double fertilization. Curr Opin Plant Biol. 2006;9:41–47. - PubMed
- Drews GN, Yadegari R. Development and function of the angiosperm female gametophyte. Annu Rev Genet. 2002;36:99–124. - PubMed
- Grossniklaus U, Schneitz K. The molecular and genetic basis of ovule and megagametophyte development. Semin Cell Dev Biol. 1998;9:227–238. - PubMed
- Higashiyama T, Yabe S, Sasaki N, Nishimura Y, Miyagishima S, et al. Pollen tube attraction by the synergid cell. Science. 2001;293:1480–1483. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases