Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing - PubMed (original) (raw)
Comparative Study
. 2007 Mar 21;26(6):1691-701.
doi: 10.1038/sj.emboj.7601603. Epub 2007 Mar 1.
Affiliations
- PMID: 17332757
- PMCID: PMC1829372
- DOI: 10.1038/sj.emboj.7601603
Comparative Study
Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing
Xianwu Zheng et al. EMBO J. 2007.
Abstract
Argonautes (AGOs) are conserved proteins that contain an RNA-binding PAZ domain and an RNase H-like PIWI domain. In Arabidopsis, except for AGO1, AGO4 and AGO7, the roles of seven other AGOs in gene silencing are not known. We found that a mutation in AGO6 partially suppresses transcriptional gene silencing in the DNA demethylase mutant ros1-1. In ago6-1ros1-1 plants, RD29A promoter short interfering RNAs (siRNAs) are less abundant, and cytosine methylation at both transgenic and endogenous RD29A promoters is reduced, compared to that in ros1-1. Interestingly, the ago4-1 mutation has a stronger suppression of the transcriptional silencing phenotype of ros1-1 mutant. Analysis of cytosine methylation at the endogenous MEA-ISR, AtREP2 and SIMPLEHAT2 loci revealed that the CpNpG and asymmetric methylation levels are lower in either of the ago6-1 and ago4-1 single mutants than those in the wild type, and the levels are the lowest in the ago6-1ago4-1 double mutant. These results suggest that AGO6 is important for the accumulation of specific heterochromatin-related siRNAs, and for DNA methylation and transcriptional gene silencing, this function is partly redundant with AGO4.
Figures
Figure 1
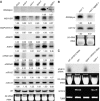
Partial suppression of RD29A-LUC and endogenous RD29A silencing but not of 35S-NPTII silencing in ros1 by ago6. (A) Luminescence image of WT, ros1-1, ros1-1ago6-1 and ago6-1. Luminescence imaging was taken with 12-day-old seedlings treated with 300 mM NaCl for 5 h. (B) Phenotype of WT, ros1-1, ros1-1ago6-1 and ago6-1 plants plated on MS medium supplemented with kanamycin (50 μg/ml). (C, D) Quantification of the luminescence intensities of WT, ros1-1, ros1-1ago6-1 and ago6-1 plants treated with 300 mM NaCl for 5 h. (E) Northern blot analysis of the transcript levels of NPT II, endogenous RD29A and the stress-responsive control gene COR15A in WT, ros1-1, ros1-1ago6-1 and ago6-1. WT, wild type of the C24 ecotype carrying a LUCIFERASE gene driven by the RD29A promoter (C24 RD29A-LUC).
Figure 2
The effect of AGO6 on DNA methylation levels. (A) DNA methylation levels (percent of methylated DNA) of transgenic RD29A promoter, and (B) DNA methylation levels of endogenous RD29A promoter in WT, ros1-1, ros1-1ago6-1 and ago6-1. (C) ago6-1 causes decreased AtSN1 cytosine methylation. PCR was used to amplify a portion of the AtSN1 retro-element. Undigested DNA and a gene lacking _Hae_III sites served as PCR controls. (D, E) DNA methylation levels at AtSN1 and 5S rDNA in WT and ago6-1.
Figure 3
Effect of ago6 on the accumulation of small RNAs. (A) Effect of ago6 on the accumulation of siRNAs from the transgenic RD29A promoter, endogenous siRNAs and miRNAs. In addition to ago6-1, a SALK T-DNA insertion allele, ago6-2 and its WT (Col-0) were also analyzed. snoRNA U6 and ethidium bromide-stained tRNA and rRNA bands served as loading controls. Relative levels of the small RNAs were calculated and shown below the small RNA bands. (B) The transgene RD29A promoter siRNA was less abundant in ros1-1ago6-1 double compared with that in the ros1-1 single mutant. (C) The effect of ago6 on the accumulation of siRNAs from transgenic inverted repeat MYB15 dsRNA. MYB15 transcript levels were analyzed by RT–PCR.
Figure 4
AGO6 gene cloning and diagram of structure. (A) AGO6 gene (At2G32940) structure. AGO6 gene has a total of 22 exons and encodes a putative AGO protein containing a PAZ domain and a PIWI domain. ago6-1 has a T-DNA insertion in the 14th exon, and ago6-2 (a T-DNA line from the SALK collection) (Salk 031553) has an insertion in the second exon. (B) Complementation of ago6-1 mutant. ros1-1ago6-1 plants transformed with WT AGO6 transgene were restored in the luminescence phenotype to that seen in ros1-1. (C) The molecular phenotype of AtSN1 methylation in ros1-1ago6-1 was restored after the ros1-1ago6-1 double mutant was transformed with the WT AGO6 gene. (D) Phylogenetic tree of the Arabidopsis AGO proteins using full-length amino-acid sequences. AGO4, 6, 8 and 9 clustered together.
Figure 5
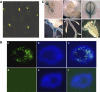
Subcellular localization of AGO6 protein and expression pattern of AGO6 promoter-driven GUS reporter gene. (A) Expression of YFP–AGO6 translational fusion under the control of CaMV 35S promoter in the epidermal cells of Arabidopsis. YFP–AGO6 protein was mainly localized in nuclei, and weakly in the cytoplasm. (B) Subcellular localization of AGO6 protein detected by immunostaining. (a) Localization of AGO6-MYC. (b) Nucleus with DAPI staining. (c) Overlap of (a) and (b). (d), (e) and (f) are negative controls showing immunostaining of a nucleus from ros1-1ago6-1 plant without the AGO6-MYC transgene. (C) AGO6 promoter–GUS expression pattern. AGO6 was strongly expressed in roots (a, e) and cotyledons (a–d), very weakly in young leaves (a, b) and not detectable in floral tissues (f).
Figure 6
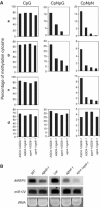
Strong suppression of RD29A-LUC and endogenous RD29A silencing in ros1-1 by ago4. (A) Luminescence images of WT, ros1-1, ros1-1ago4-1 and ago4-1 seedlings. Luminescence images were taken with 12-day-old seedlings treated with 300 mM NaCl for 5 h. (B) Northern blot analysis of the transcript levels of endogenous RD29A and the stress-responsive control gene COR15A in WT, ros1-1, ros1-1ago4-1 and ago4-1. (C) DNA methylation levels (percentage of methylated cytosine) of transgenic RD29A promoter, and (D) DNA methylation levels of endogenous RD29A promoter in WT, ros1-1 and ros1-1ago4-1.
Figure 7
DNA methylation analysis at several endogenous loci in AGO4-1AGO6-1, AGO4-1ago6-1, ago4-1AGO6-1 and ago4-1ago6-1. (A) CpG (left), CpNpG (middle) and CpNpN (right) methylation at the MEA-ISR (a), AtREP2 (b), SIMPLEHAT2 (c) and AtGP1 (d) loci were analyzed by bisulfite sequencing. Methylation levels are shown by the percentage of methylated cytosine in all sequenced clones. Detailed bisulfite sequencing data are in Supplementary Figures. (B) Northern blot analysis of siRNAs at the AtREP2 locus in ago4-1, ago6-1 single mutants and ago4-1ago6-1 double mutant. miR172 was used as a control.
Similar articles
- Specific but interdependent functions for Arabidopsis AGO4 and AGO6 in RNA-directed DNA methylation.
Duan CG, Zhang H, Tang K, Zhu X, Qian W, Hou YJ, Wang B, Lang Z, Zhao Y, Wang X, Wang P, Zhou J, Liang G, Liu N, Wang C, Zhu JK. Duan CG, et al. EMBO J. 2015 Mar 4;34(5):581-92. doi: 10.15252/embj.201489453. Epub 2014 Dec 19. EMBO J. 2015. PMID: 25527293 Free PMC article. - Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA-methylation-independent manner in Arabidopsis.
Kapoor A, Agarwal M, Andreucci A, Zheng X, Gong Z, Hasegawa PM, Bressan RA, Zhu JK. Kapoor A, et al. Curr Biol. 2005 Nov 8;15(21):1912-8. doi: 10.1016/j.cub.2005.09.013. Curr Biol. 2005. PMID: 16271867 - ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation.
Zilberman D, Cao X, Jacobsen SE. Zilberman D, et al. Science. 2003 Jan 31;299(5607):716-9. doi: 10.1126/science.1079695. Epub 2003 Jan 9. Science. 2003. PMID: 12522258 - Preventing transcriptional gene silencing by active DNA demethylation.
Kapoor A, Agius F, Zhu JK. Kapoor A, et al. FEBS Lett. 2005 Oct 31;579(26):5889-98. doi: 10.1016/j.febslet.2005.08.039. Epub 2005 Aug 31. FEBS Lett. 2005. PMID: 16162337 Review. - Epigenetic silencing of endogenous repetitive sequences by MORPHEUS' MOLECULE1 in Arabidopsis thaliana.
Habu Y. Habu Y. Epigenetics. 2010 Oct 1;5(7):562-5. doi: 10.4161/epi.5.7.12518. Epigenetics. 2010. PMID: 21464619 Review.
Cited by
- Transgenerational maintenance of transgene body CG but not CHG and CHH methylation.
Dalakouras A, Dadami E, Zwiebel M, Krczal G, Wassenegger M. Dalakouras A, et al. Epigenetics. 2012 Sep;7(9):1071-8. doi: 10.4161/epi.21644. Epub 2012 Aug 6. Epigenetics. 2012. PMID: 22863736 Free PMC article. - The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications.
Tang K, Lang Z, Zhang H, Zhu JK. Tang K, et al. Nat Plants. 2016 Oct 31;2(11):16169. doi: 10.1038/nplants.2016.169. Nat Plants. 2016. PMID: 27797352 Free PMC article. - The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana.
Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW. Wang XB, et al. Plant Cell. 2011 Apr;23(4):1625-38. doi: 10.1105/tpc.110.082305. Epub 2011 Apr 5. Plant Cell. 2011. PMID: 21467580 Free PMC article. - Two domain-disrupted hda6 alleles have opposite epigenetic effects on transgenes and some endogenous targets.
Zhang S, Zhan X, Xu X, Cui P, Zhu JK, Xia Y, Xiong L. Zhang S, et al. Sci Rep. 2015 Dec 15;5:17832. doi: 10.1038/srep17832. Sci Rep. 2015. PMID: 26666962 Free PMC article. - RNA-directed DNA methylation and demethylation in plants.
Chinnusamy V, Zhu JK. Chinnusamy V, et al. Sci China C Life Sci. 2009 Apr;52(4):331-43. doi: 10.1007/s11427-009-0052-1. Epub 2009 Apr 21. Sci China C Life Sci. 2009. PMID: 19381459 Free PMC article. Review.
References
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 - PubMed
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3-type myb transcription factor is involved in the cold-regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281: 37636–37645 - PubMed
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 - PubMed
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases