Meta-analysis in genome-wide association datasets: strategies and application in Parkinson disease - PubMed (original) (raw)
Meta-Analysis
Meta-analysis in genome-wide association datasets: strategies and application in Parkinson disease
Evangelos Evangelou et al. PLoS One. 2007.
Abstract
Background: Genome-wide association studies hold substantial promise for identifying common genetic variants that regulate susceptibility to complex diseases. However, for the detection of small genetic effects, single studies may be underpowered. Power may be improved by combining genome-wide datasets with meta-analytic techniques.
Methodology/principal findings: Both single and two-stage genome-wide data may be combined and there are several possible strategies. In the two-stage framework, we considered the options of (1) enhancement of replication data and (2) enhancement of first-stage data, and then, we also considered (3) joint meta-analyses including all first-stage and second-stage data. These strategies were examined empirically using data from two genome-wide association studies (three datasets) on Parkinson disease. In the three strategies, we derived 12, 5, and 49 single nucleotide polymorphisms that show significant associations at conventional levels of statistical significance. None of these remained significant after conservative adjustment for the number of performed analyses in each strategy. However, some may warrant further consideration: 6 SNPs were identified with at least 2 of the 3 strategies and 3 SNPs [rs1000291 on chromosome 3, rs2241743 on chromosome 4 and rs3018626 on chromosome 11] were identified with all 3 strategies and had no or minimal between-dataset heterogeneity (I(2) = 0, 0 and 15%, respectively). Analyses were primarily limited by the suboptimal overlap of tested polymorphisms across different datasets (e.g., only 31,192 shared polymorphisms between the two tier 1 datasets).
Conclusions/significance: Meta-analysis may be used to improve the power and examine the between-dataset heterogeneity of genome-wide association studies. Prospective designs may be most efficient, if they try to maximize the overlap of genotyping platforms and anticipate the combination of data across many genome-wide association studies.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
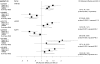
Figure 1
Meta-analyses of the three datasets for the 6 single nucleotide polymorphisms that were selected (p<0.05 unadjusted for multiple comparisons) with at least two of the three strategies. For each polymorphism the forest plot shows the odds ratio and 95% confidence interval for each dataset as well as the summary odds ratio and 95% confidence intervals by random effects calculations. Also shown is the p-value for the summary effect and the I-squared statistic for between-dataset heterogeneity.
Similar articles
- Heterogeneity in meta-analyses of genome-wide association investigations.
Ioannidis JP, Patsopoulos NA, Evangelou E. Ioannidis JP, et al. PLoS One. 2007 Sep 5;2(9):e841. doi: 10.1371/journal.pone.0000841. PLoS One. 2007. PMID: 17786212 Free PMC article. - Meta-analysis of genetic association studies: methodologies, between-study heterogeneity and winner's curse.
Nakaoka H, Inoue I. Nakaoka H, et al. J Hum Genet. 2009 Nov;54(11):615-23. doi: 10.1038/jhg.2009.95. Epub 2009 Oct 23. J Hum Genet. 2009. PMID: 19851339 Review. - Meta-analysis of genome-wide association studies.
de Bakker PI, Neale BM, Daly MJ. de Bakker PI, et al. Cold Spring Harb Protoc. 2010 Jun;2010(6):pdb.top81. doi: 10.1101/pdb.top81. Cold Spring Harb Protoc. 2010. PMID: 20516189 - Genome-wide association study confirms BST1 and suggests a locus on 12q24 as the risk loci for Parkinson's disease in the European population.
Saad M, Lesage S, Saint-Pierre A, Corvol JC, Zelenika D, Lambert JC, Vidailhet M, Mellick GD, Lohmann E, Durif F, Pollak P, Damier P, Tison F, Silburn PA, Tzourio C, Forlani S, Loriot MA, Giroud M, Helmer C, Portet F, Amouyel P, Lathrop M, Elbaz A, Durr A, Martinez M, Brice A; French Parkinson's Disease Genetics Study Group. Saad M, et al. Hum Mol Genet. 2011 Feb 1;20(3):615-27. doi: 10.1093/hmg/ddq497. Epub 2010 Nov 17. Hum Mol Genet. 2011. PMID: 21084426 - The pursuit of genome-wide association studies: where are we now?
Ku CS, Loy EY, Pawitan Y, Chia KS. Ku CS, et al. J Hum Genet. 2010 Apr;55(4):195-206. doi: 10.1038/jhg.2010.19. Epub 2010 Mar 19. J Hum Genet. 2010. PMID: 20300123 Review.
Cited by
- Quick, "imputation-free" meta-analysis with proxy-SNPs.
Meesters C, Leber M, Herold C, Angisch M, Mattheisen M, Drichel D, Lacour A, Becker T. Meesters C, et al. BMC Bioinformatics. 2012 Sep 12;13:231. doi: 10.1186/1471-2105-13-231. BMC Bioinformatics. 2012. PMID: 22971100 Free PMC article. - An integrative genomics approach to biomarker discovery in breast cancer.
Hicks C, Asfour R, Pannuti A, Miele L. Hicks C, et al. Cancer Inform. 2011;10:185-204. doi: 10.4137/CIN.S6837. Epub 2011 Jul 25. Cancer Inform. 2011. PMID: 21869864 Free PMC article. - The impact of imputation on meta-analysis of genome-wide association studies.
Li J, Guo YF, Pei Y, Deng HW. Li J, et al. PLoS One. 2012;7(4):e34486. doi: 10.1371/journal.pone.0034486. Epub 2012 Apr 5. PLoS One. 2012. PMID: 22496814 Free PMC article. - A genomic pathway approach to a complex disease: axon guidance and Parkinson disease.
Lesnick TG, Papapetropoulos S, Mash DC, Ffrench-Mullen J, Shehadeh L, de Andrade M, Henley JR, Rocca WA, Ahlskog JE, Maraganore DM. Lesnick TG, et al. PLoS Genet. 2007 Jun;3(6):e98. doi: 10.1371/journal.pgen.0030098. PLoS Genet. 2007. PMID: 17571925 Free PMC article. - Robust meta-analysis for large-scale genomic experiments based on an empirical approach.
Sikdar S. Sikdar S. BMC Med Res Methodol. 2022 Feb 10;22(1):43. doi: 10.1186/s12874-022-01530-y. BMC Med Res Methodol. 2022. PMID: 35144554 Free PMC article.
References
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. - PubMed
- Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109–18. - PubMed
- Todd JA. Statistical false positive or true disease pathway? Nat Genet. 2006;38:731–3. - PubMed
- Ioannidis JP. Genetic associations: false or true? Trends Mol Med. 2003;9:135–8. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials