Visualization of three-way comparisons of omics data - PubMed (original) (raw)
Comparative Study
Visualization of three-way comparisons of omics data
Richard Baran et al. BMC Bioinformatics. 2007.
Abstract
Background: Density plot visualizations (also referred to as heat maps or color maps) are widely used in different fields including large-scale omics studies in biological sciences. However, the current color-codings limit the visualizations to single datasets or pairwise comparisons.
Results: We propose a color-coding approach for the representation of three-way comparisons. The approach is based on the HSB (hue, saturation, brightness) color model. The three compared values are assigned specific hue values from the circular hue range (e.g. red, green, and blue). The hue value representing the three-way comparison is calculated according to the distribution of three compared values. If two of the values are identical and one is different, the resulting hue is set to the characteristic hue of the differing value. If all three compared values are different, the resulting hue is selected from a color gradient running between the hues of the two most distant values (as measured by the absolute value of their difference) according to the relative position of the third value between the two. The saturation of the color representing the three-way comparison reflects the amplitude (or extent) of the numerical difference between the two most distant values according to a scale of interest. The brightness is set to a maximum value by default but can be used to encode additional information about the three-way comparison.
Conclusion: We propose a novel color-coding approach for intuitive visualization of three-way comparisons of omics data.
Figures
Figure 1
Examples of color-codings for three-way comparisons. Color representations for three-way comparisons of selected values a, b, and c calculated using the proposed procedure are shown in the column labeled HSB-based. Colors acquired by substituting values of a, b, and c directly for red, green, and blue or cyan, magenta, and yellow (black = 0) are shown in columns labeled RGB or CMYK, respectively. The legend is drawn as a hexagon instead of a circle for convenience. Horizontal lines separate groups of values with similar distributions.
Figure 2
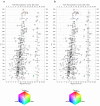
Metabolite profiles for the three-way comparison. Mouse liver extract metabolite profiles acquired by CE-TOFMS two hours after intraperitoneal injection with (a) vehicle (Control), (b) diethylmaleate (DEM), a non-protein thiol-depleting chemical, or (c) buthionine sulfoximine (BSO), an inhibitor of γ-glutamylcysteine synthase. The plotted datasets are averages of five normalized replicate datasets for cation measurements originating from our previous work [2]. The averaged datasets are visualized as density plots. For all plots, numbered ovals (annotation labels) indicate the expected locations of peaks of a set of known chemical compounds and are used for identification of metabolites on the density plots [2,3].
Figure 3
Three-way comparison of metabolite profiles. (a) Absolute × relative three-way comparison of metabolite profiles shown in Figure 2. Averages of replicate datasets (n = 5) were used for the three-way comparison. The resulting dataset was filtered using F-ratio (one-way ANOVA) to select only statistically significant differences as described in the main text. (b) The Control dataset (Figure 2a) was overlaid on the three-way comparison result shown in panel (a) via the brightness value. Darkening of the colored spots indicates the size of the corresponding peaks in the Control dataset. Gray spots show peaks which do not significantly differ among the datasets. For both plots, numbered ovals (annotation labels) indicate the expected locations of peaks of a set of known chemical compounds and are used for identification of metabolites on the density plots [2,3].
Figure 4
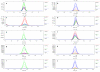
Candidate differences. Overlaid extracted ion electropherograms for the most significant differences from the three-way comparison results shown in Figure 3. Each panel represents data in the form of signal intensity (number of ions) over time for a specific mass interval (1 Da bin). The vertical dashed line indicates the position of the most significant difference according to the three-way comparison results. When present within panels, numbers correspond to the annotation labels in Figures 2 and 3.
Similar articles
- Color distribution of a shade guide in the value, chroma, and hue scale.
Ahn JS, Lee YK. Ahn JS, et al. J Prosthet Dent. 2008 Jul;100(1):18-28. doi: 10.1016/S0022-3913(08)60129-8. J Prosthet Dent. 2008. PMID: 18589070 - Weaving versus blending: a quantitative assessment of the information carrying capacities of two alternative methods for conveying multivariate data with color.
Hagh-Shenas H, Kim S, Interrante V, Healey C. Hagh-Shenas H, et al. IEEE Trans Vis Comput Graph. 2007 Nov-Dec;13(6):1270-7. doi: 10.1109/TVCG.2007.70623. IEEE Trans Vis Comput Graph. 2007. PMID: 17968074 - 2D molecular graphics: a flattened world of chemistry and biology.
Zhou P, Shang Z. Zhou P, et al. Brief Bioinform. 2009 May;10(3):247-58. doi: 10.1093/bib/bbp013. Epub 2009 Mar 30. Brief Bioinform. 2009. PMID: 19332474 - Integration of omics data: how well does it work for bacteria?
De Keersmaecker SC, Thijs IM, Vanderleyden J, Marchal K. De Keersmaecker SC, et al. Mol Microbiol. 2006 Dec;62(5):1239-50. doi: 10.1111/j.1365-2958.2006.05453.x. Epub 2006 Oct 16. Mol Microbiol. 2006. PMID: 17040488 Review. - The model organism as a system: integrating 'omics' data sets.
Joyce AR, Palsson BØ. Joyce AR, et al. Nat Rev Mol Cell Biol. 2006 Mar;7(3):198-210. doi: 10.1038/nrm1857. Nat Rev Mol Cell Biol. 2006. PMID: 16496022 Review.
Cited by
- Circos: an information aesthetic for comparative genomics.
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Krzywinski M, et al. Genome Res. 2009 Sep;19(9):1639-45. doi: 10.1101/gr.092759.109. Epub 2009 Jun 18. Genome Res. 2009. PMID: 19541911 Free PMC article. - Metabolites associated with adaptation of microorganisms to an acidophilic, metal-rich environment identified by stable-isotope-enabled metabolomics.
Mosier AC, Justice NB, Bowen BP, Baran R, Thomas BC, Northen TR, Banfield JF. Mosier AC, et al. mBio. 2013 Mar 12;4(2):e00484-12. doi: 10.1128/mBio.00484-12. mBio. 2013. PMID: 23481603 Free PMC article. - Cocultivation of Anaerobic Fungi with Rumen Bacteria Establishes an Antagonistic Relationship.
Swift CL, Louie KB, Bowen BP, Hooker CA, Solomon KV, Singan V, Daum C, Pennacchio CP, Barry K, Shutthanandan V, Evans JE, Grigoriev IV, Northen TR, O'Malley MA. Swift CL, et al. mBio. 2021 Aug 31;12(4):e0144221. doi: 10.1128/mBio.01442-21. Epub 2021 Aug 17. mBio. 2021. PMID: 34399620 Free PMC article. - VennPainter: A Tool for the Comparison and Identification of Candidate Genes Based on Venn Diagrams.
Lin G, Chai J, Yuan S, Mai C, Cai L, Murphy RW, Zhou W, Luo J. Lin G, et al. PLoS One. 2016 Apr 27;11(4):e0154315. doi: 10.1371/journal.pone.0154315. eCollection 2016. PLoS One. 2016. PMID: 27120465 Free PMC article.
References
- Soga T, Baran R, Suematsu M, Ueno Y, Ikeda S, Sakurakawa T, Kakazu Y, Ishikawa T, Robert M, Nishioka T, Tomita M. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J Biol Chem. 2006;281:16768–16776. doi: 10.1074/jbc.M601876200. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources