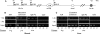Analysis of the H19ICR insulator - PubMed (original) (raw)
Analysis of the H19ICR insulator
Young Soo Yoon et al. Mol Cell Biol. 2007 May.
Abstract
Transcriptional insulators are specialized cis-acting elements that protect promoters from inappropriate activation by distal enhancers. The H19 imprinting control region (ICR) functions as a CTCF-dependent, methylation-sensitive transcriptional insulator. We analyzed several insertional mutations and demonstrate that the ICR can function as a methylation-regulated maternal chromosome-specific insulator in novel chromosomal contexts. We used chromosome conformation capture and chromatin immunoprecipitation assays to investigate the configuration of cis-acting elements at these several insertion sites. By comparing maternal and paternal organizations on wild-type and mutant chromosomes, we hoped to identify mechanisms for ICR insulator function. We found that promoter and enhancer elements invariably associate to form DNA loop domains at transcriptionally active loci. Conversely, active insulators always prevent these promoter-enhancer interactions. Instead, the ICR insulator forms novel loop domains by associating with the blocked promoters and enhancers. We propose that these associations are fundamental to insulator function.
Figures
FIG. 1.
Long-range interactions at the Igf2 locus on wild-type (WT) and Δ_ICR_ chromosomes. (A) Schematic depiction of the 100-kb Igf2_-H19 locus includes the three Igf2 promoters (P1 at kb −78, P2 at kb −76, and P3 at kb −74), the shared ICR (at kb −4.4 to −2), the H19 promoter (H19P at bp 0), and the shared endodermal (open circle at kb +8) and mesodermal (filled circle at kb +25) enhancers. DMR1 and DMR2, flanking the Igf2 promoters, become methylated on the paternal chromosome in the postimplantation embryo and play a role in the activation of paternal Igf2 in liver cells and in the repression of maternal Igf2 in muscle cells, respectively. The Δ_ICR chromosome carries a 5-kb deletion from kb −6 to −1 that removes the ICR. The vertical bars above and below the map indicate BamHI and BglII restriction sites, respectively. Arrowheads depict the orientations and locations of PCR primers used for 3C analysis. Asterisks indicate RFLPs that distinguish between wild-type M. castaneus alleles and M. domesticus alleles. (B to K) 3C analysis of long-range interactions at the Igf2 locus was carried out on using the primers indicated. Animal genotypes C/D, D/C, C/Δ_ICR_, and Δ_ICR_/C are indicated (maternal allele listed first). The top panels for each experiment represent the 3C PCR product. The bottom panels, when included, depict the banding patterns after digestion with enzymes distinguishing between the M. castaneus (C-labeled arrowheads)- and M. domesticus (D-labeled arrowheads)-derived DNAs. Note that the Δ_ICR_ mutation is on an M. domesticus chromosome. (B and C) In wild-type muscle cells (B) and liver cells (C), the Igf2 promoters associate with the mesodermal and endodermal enhancers, respectively, only on the active paternal chromosome. That is, the C/D and D/C extracts yield only PCR products that are all M. domesticus (D-labeled arrowheads) and all M. castaneus (C-labeled arrowheads) alleles, respectively. (D and E) The maternally inherited ICR insulator is necessary to prevent maternal promoter-enhancer interactions in both muscle (D) and liver (E) cells. Maternal inheritance of the ICR deletion mutant results in biallelic interactions between the Igf2 promoters and the enhancers, as indicated by the presence of both M. domesticus and M. castaneus bands in the 3C products of Δ_ICR_/C extracts. (F and G) Inactive maternal Igf2 promoters associate with the ICR insulator in both muscle (F) and liver (G) cells. (H and I) Igf2 promoters interact only with a maternally inherited ICR in both muscle (H) and liver (I) cells. (J) The maternal ICR interacts with blocked downstream enhancers. (K) Only the unmethylated maternal ICR interacts with downstream mesodermal and endodermal enhancers. NS, nonspecific PCR product; C+D, digestion products indicative of both M. castaneus and M. domesticus DNAs comigrate.
FIG. 2.
Long-range interactions at the H19 locus on wild-type and H19R chromosomes. (A) Schematic representation of the _Igf2_-H19 locus, including the Igf2 promoter 1 (Igf2 P1 at kb −78), the ICR (at kb −4.4 to −2), the H19 promoter (H19 P at bp 0), and the shared endodermal (open circle at kb 8) and mesodermal (filled circle at kb 25) enhancers. The H19R mutation, depicted on the lower line, is an insertion of the 2.4-kb ICR fragment at the kb +10 EcoRI site. The vertical bars above and below the maps indicate BamHI and BglII restriction sites, respectively. Arrowheads depict the orientations and locations of PCR primers used for 3C and ChIP analysis. Asterisks indicate RFLPs that distinguish between M. castaneus and M. domesticus alleles. (B) ChIP analyses demonstrate that CTCF proteins can bind in vivo to the ICR insertion on H19R. After preparing cross-linked protein-DNA extracts, the presence of the endogenous and H19R ICR sequences was detected by PCR amplification for 45, 43, or 41 cycles with the primers indicated. Ab, antisera. (C) The 2.4-kb ICR element is a transcriptional insulator at a heterologous location. RNAs prepared from liver (left panel) and muscle (middle and right panels) of P2 littermates were analyzed by Northern blotting using probes specific to H19 or Igf2, as indicated. Subsequently, blots were stripped and hybridized with probes to Elongation Factor 2 (EF2). +/+, wild-type maternal and paternal chromosomes; H19R/+, maternal inheritance of the H19R chromosome; +/H19R, paternal inheritance of the H19R chromosome. (D) In wild-type muscle cells and liver cells, the H19 promoter associates with the mesodermal and endodermal enhancers, respectively, only on the active maternal chromosome. 3C analysis was performed using the primers indicated and extracts from wild-type C/D and D/C pups. Top panels depict the 3C PCR product. Bottom panels depict the banding patterns after digestion with enzymes distinguishing the M. castaneus (C-labeled arrowheads)- and M. domesticus (D-labeled arrowheads)-derived DNAs. (E) The H19R ICR insertion blocks H19 promoter-enhancer associations in muscle but not in liver. 3C analysis was performed on extracts prepared from liver and muscle cells from H19R/C and D/C animals. Primers 9 plus 16 test for H19 promoter-endodermal enhancer interactions, while primers 12 plus 17 test for H19 promoter-mesodermal enhancer interactions. Primer pairs 2 plus 9 and 2 plus 13 identify paternal Igf2 promoter-enhancer interactions (endodermal and mesodermal, respectively) and control for the integrity of the extracts. Each PCR was analyzed at 42, 40, and 38 cycles.
FIG. 3.
Long-range interactions on the wild-type and AfpICR chromosomes. (A) Schematic representation of the wild-type Afp locus (top line) and the ICR insertion mutation on AfpICR (bottom line). The three enhancer elements (E3, E2, and E1) and the Afp promoter (AfpP, horizontal arrow) are depicted. Vertical lines above and below the maps represent BamHI and BglII sites, respectively. Arrowheads depict the orientations and locations of PCR primers used for 3C and ChIP. Asterisks indicate RFLPs that distinguish between M. castaneus and M. domesticus alleles. Note that the AfpICR insertion is on an M. domesticus chromosome. (B) Maternal chromosome-specific binding of CTCF to the AfpICR insertion. After cross-linked protein-DNA extracts were prepared, the presence of ICR sequences was detected by real-time quantitative PCR using the primers ICR-F and ICR-R. The amount of ICR DNA was determined for samples treated with (+ Ab) or without (− Ab) antisera to CTCF and then compared to input (genomic) DNA, and that ratio is reported. For comparison, all samples were also analyzed using primers specific for the CTCF binding sites at the β-globin locus (LCR). (C) 3C analysis was performed using the primers 46 plus 51 and liver extracts prepared from D/C, C/D, and AfpICR/C animals. C-labeled arrowhead, M. castaneus allele product; D-labeled arrowhead, M. domesticus allele product.
FIG. 4.
ChIP assays confirm the proximity of the insulator and promoter and enhancer elements on the maternal chromosome. (A) Schematic representation of the _Igf2_-H19 locus including the three Igf2 promoters, the ICR, the H19 promoter, and the shared endodermal (open circle) and mesodermal (closed circle) enhancers. Numbers above the line indicate the relative positions of the corresponding elements. Arrowheads indicate the locations and orientations of primers used the ChIP analysis. (B and C) Cross-linked extracts were prepared from muscle (B) and liver (C) cells and analyzed by ChIP using polyclonal antisera specific to CTCF protein. ICR, enhancer, and Igf2 promoter 2 sequences were identified using the primers indicated and quantitated by testing PCR products after 41, 43, and 45 cycles. In addition, primers specific to the CD3 locus were tested as a nonspecific control. + Ab, with antisera; − Ab, without antisera.
Similar articles
- Three-dimensional conformation at the H19/Igf2 locus supports a model of enhancer tracking.
Engel N, Raval AK, Thorvaldsen JL, Bartolomei SM. Engel N, et al. Hum Mol Genet. 2008 Oct 1;17(19):3021-9. doi: 10.1093/hmg/ddn200. Epub 2008 Jul 10. Hum Mol Genet. 2008. PMID: 18617529 Free PMC article. - CTCF-dependent chromatin insulator is linked to epigenetic remodeling.
Ishihara K, Oshimura M, Nakao M. Ishihara K, et al. Mol Cell. 2006 Sep 1;23(5):733-42. doi: 10.1016/j.molcel.2006.08.008. Mol Cell. 2006. PMID: 16949368 - Influence of a CTCF-Dependent Insulator on Multiple Aspects of Enhancer-Mediated Chromatin Organization.
Varma G, Rawat P, Jalan M, Vinayak M, Srivastava M. Varma G, et al. Mol Cell Biol. 2015 Oct;35(20):3504-16. doi: 10.1128/MCB.00514-15. Epub 2015 Aug 3. Mol Cell Biol. 2015. PMID: 26240285 Free PMC article. - [The progress in the study of chromatin insulator].
Zhang B, Liu X. Zhang B, et al. Yi Chuan. 2004 Jul;26(4):551-5. Yi Chuan. 2004. PMID: 15640060 Review. Chinese. - Chromatin insulators and long-distance interactions in Drosophila.
Kyrchanova O, Georgiev P. Kyrchanova O, et al. FEBS Lett. 2014 Jan 3;588(1):8-14. doi: 10.1016/j.febslet.2013.10.039. Epub 2013 Nov 5. FEBS Lett. 2014. PMID: 24211836 Review.
Cited by
- The Igf2/H19 muscle enhancer is an active transcriptional complex.
Eun B, Sampley ML, Van Winkle MT, Good AL, Kachman MM, Pfeifer K. Eun B, et al. Nucleic Acids Res. 2013 Sep;41(17):8126-34. doi: 10.1093/nar/gkt597. Epub 2013 Jul 10. Nucleic Acids Res. 2013. PMID: 23842673 Free PMC article. - Chromatin insulators: linking genome organization to cellular function.
Phillips-Cremins JE, Corces VG. Phillips-Cremins JE, et al. Mol Cell. 2013 May 23;50(4):461-74. doi: 10.1016/j.molcel.2013.04.018. Mol Cell. 2013. PMID: 23706817 Free PMC article. Review. - CTCF-dependent enhancer-blocking by alternative chromatin loop formation.
Hou C, Zhao H, Tanimoto K, Dean A. Hou C, et al. Proc Natl Acad Sci U S A. 2008 Dec 23;105(51):20398-403. doi: 10.1073/pnas.0808506106. Epub 2008 Dec 12. Proc Natl Acad Sci U S A. 2008. PMID: 19074263 Free PMC article. - A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber.
Comet I, Schuettengruber B, Sexton T, Cavalli G. Comet I, et al. Proc Natl Acad Sci U S A. 2011 Feb 8;108(6):2294-9. doi: 10.1073/pnas.1002059108. Epub 2011 Jan 24. Proc Natl Acad Sci U S A. 2011. PMID: 21262819 Free PMC article. - Cardiac pathologies in mouse loss of imprinting models are due to misexpression of H19 long noncoding RNA.
Park KS, Rahat B, Lee HC, Yu ZX, Noeker J, Mitra A, Kean CM, Knutsen RH, Springer D, Gebert CM, Kozel BA, Pfeifer K. Park KS, et al. Elife. 2021 Aug 17;10:e67250. doi: 10.7554/eLife.67250. Elife. 2021. PMID: 34402430 Free PMC article.
References
- Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7:1663-1673. - PubMed
- Bartolomei, M. S., S. Zemel, and S. M. Tilghman. 1991. Parental imprinting of the mouse H19 gene. Nature 351:153-155. - PubMed
- Bell, A., and G. Felsenfeld. 1999. Stopped at the border: boundaries and insulators. Curr. Opin. Genet. Dev. 9:191-198. - PubMed
- Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. - PubMed
- Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases