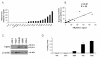Heparanase influences expression and shedding of syndecan-1, and its expression by the bone marrow environment is a bad prognostic factor in multiple myeloma - PubMed (original) (raw)
. 2007 Jun 1;109(11):4914-23.
doi: 10.1182/blood-2006-08-043232. Epub 2007 Mar 5.
Dirk Hose, Pierre Raynaud, Michael Hundemer, Michel Jourdan, Eric Jourdan, Veronique Pantesco, Marion Baudard, John De Vos, Marion Larroque, Thomas Moehler, Jean-Francois Rossi, Thierry Rème, Hartmut Goldschmidt, Bernard Klein
Affiliations
- PMID: 17339423
- PMCID: PMC2268882
- DOI: 10.1182/blood-2006-08-043232
Heparanase influences expression and shedding of syndecan-1, and its expression by the bone marrow environment is a bad prognostic factor in multiple myeloma
Karène Mahtouk et al. Blood. 2007.
Abstract
The heparan sulfate (HS) proteoglycan, syndecan-1, plays a major role in multiple myeloma (MM) by concentrating heparin-binding growth factors on the surface of MM cells (MMCs). Using Affymetrix microarrays and real-time reverse transcriptase-polymerase chain reaction (RT-PCR), we show that the gene encoding heparanase (HPSE), an enzyme that cleaves HS chains, is expressed by 11 of 19 myeloma cell lines (HMCLs). In HSPE(pos) HMCLs, syndecan-1 gene expression and production of soluble syndecan-1, unlike expression of membrane syndecan-1, were significantly increased. Knockdown of HPSE by siRNA resulted in a decrease of syndecan-1 gene expression and soluble syndecan-1 production without affecting membrane syndecan-1 expression. Thus, HPSE influences expression and shedding of syndecan-1. Contrary to HMCLs, HPSE is expressed in only 4 of 39 primary MMC samples, whereas it is expressed in 36 of 39 bone marrow (BM) microenvironment samples. In the latter, HPSE is expressed at a median level in polymorphonuclear cells and T cells; it is highly expressed in monocytes and osteoclasts. Affymetrix data were validated at the protein level, both on HMCLs and patient samples. We report for the first time that a gene's expression mainly in the BM environment (ie, HSPE) is associated with a shorter event-free survival of patients with newly diagnosed myeloma treated with high-dose chemotherapy and stem cell transplantation. Our study suggests that clinical inhibitors of HPSE could be beneficial for patients with MM.
Figures
Figure 1. HPSE expression and activity are heterogeneous in HMCLs
A. Expression of HPSE in 19 HMCLs determined using Affymetrix U133 set microarrays. White and black histograms indicate that the Affymetrix call is “absent” or “present”, respectively. B. Correlation between Affymetrix and real-time RT-PCR HPSE expression data. For real-time RT-PCR, HPSE expression in each sample was normalized to that of GAPDH and the XG-2 cell line was used as a reference with the arbitrary value of 100. C. HMCLs were lysed and the lysates were separated on a 12% SDS-PAGE and analyzed by Western blot with a polyclonal anti-HPSE antibody. Both the 65 kDa MW pro-enzyme and the 50 kDa MW active form were identified. β-actin was used as a loading control. Results are of one experiment representative of three. kDa, molecular weight in thousands. D. HPSE activity was determined using an ELISA-type detection assay. HMCLs lysates were incubated with biotinylated HS (b-HS) and then only undegraded b-HS could bind an FGF-coated ELISA plate. Bound b-HS was detected with HRP-streptavidin followed by a colorimetric assay. HPSE activity corresponding to absorbance at 450 nm was determined by comparison with a standard curve as described in “Material and Methods”. One unit is defined as the activity that can degrade 0.063 ng of b-HS when reacted at pH 5.8 at 37°C for one min. The detection limit (dotted line) was 0.05 U/ml. Results are of one experiment representative of three.
Figure 2. HMCLs that express HPSE produce higher amounts of soluble syndecan-1
A. Syndecan-1 expression was determined in the 19 HMCLs using Affymetrix U133 set microarrays. B. Mean fluorescence intensity of membrane syndecan-1 expression determined with a PE-conjugated anti-GDI38 MoAb, a PE-isotype-matched control MoAb, and FACS analysis. For each cell line, the mean fluorescence intensity obtained with the control MoAb was set between 4–6. C. For the 19 HMCLs, the rates of production of soluble syndecan-1 per 106 cells and per 24 h was determined during the 2nd and 3rd days of culture (exponential growth phase) and measured by ELISA. The dotted line indicates the detection limit of the test (<8 ng/mL). D. For XG-2 and U266 HMCLs, the cell count, the density of membrane syndecan-1 and the concentration of soluble syndecan-1 in the culture supernatant were determined each day for three days.
Figure 3. Syndecan-1 mRNA and soluble syndecan-1 are downregulated in _HPSE_-silenced cells
U266 cells were electroporated with no siRNA or with a non-targeting control siRNA (siRNA-co) or with an _HPSE_-specific siRNA (siRNA-HPSE), and cultured for two days. A. At day 2, HPSE, syndecan-1, and GAPDH expression were quantified by real-time RT-PCR and were normalized for each sample to that of B2M. Cells electroporated with no siRNA were used as a reference and were assigned the arbitrary value of 100. Data are means ±SD of the gene expression levels determined for five independent experiments. *Indicates that the mean value is statistically significantly from that obtained with the control (no siRNA), using a Student t test for pairs (P≤.05). B. Membrane expression of syndecan-1 was determined with a PE-conjugated anti-GDI38 MoAb and a PE-isotype-matched control MoAb and FACS analysis. Results shown are those of one experiment representative of five. C. U266 cells were electroporated with no siRNA or with a non-targeting control siRNA (siRNA-co) or with an _HPSE_-specific siRNA (siRNA-HPSE). Data are expressed as the means ±SD of the rates of production of soluble syndecan-1 per 106 cells and per 24 h determined during the first and second days of culture after electroporation determined in five separate experiments. Indicates that the mean value is statistically significantly different from that obtained in the control (no siRNA), using a Student f test for pairs (P≤.05).
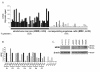
Figure 4. Comparison of HPSE expression between WBM and the corresponding purified MMC of patients
A. Expression of HPSE was determined in the WBM of 39 myeloma patients as well as in the myeloma cells purified from the bone marrow of the same patients, using Affymetrix U1332 Plus 2.0 microarrays. White histograms indicate that the Affymetrix call is “absent”; black (WBM) and grey (MMC) histograms indicate that it is “present”. B. HPSE expression was measured by real-time RT-PCR in the whole bone marrow of seven MM patients (WBM, black bars), in the corresponding MMC (grey bars), in the corresponding microenvironment depleted from myeloma cells (<2% MMC, ENV, crosshatched bars) and in MMC from five patients with PCL. HPSE expression was normalized to that of GAPDH. For each patient, the WBM sample was used as a reference and was assigned the arbitrary value of 100. For the five patients with PCL, the WBM sample of patient 1 was used as a reference. The median percentage of plasma cells in bone marrow aspirates from the seven patients with intramedullary myeloma was 8,5% (range=6–40). C. HPSE protein expression was determined in microenvironment cells depleted from MMC (< 2% MMC, ENV1-5) of five patients and in purified MMC of three other patients (MMC1-3). Cells were lysed and the lysates were separated on a 12% SDS-PAGE and analyzed by Western blot with a polyclonal anti-HPSE antibody. Both the 65 kDa MW pro-enzyme and the 50 kDa MW active form were identified. β-actin was used as a loading control. Results are from two separate western blots, one with 3 purified MMC, the other one with the 5 microenvironment cells. The HPSEpos XG-2 HMCL was used as a positive HPSE protein control in the two blots (results not shown). kDa, molecular weight in thousands.
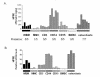
Figure 5. Expression of HPSE in subpopulations of the BM environment of patients with MM
A. Expression of HPSE was determined in the WBM of five MM patients as well as in MMC, CD3+ cells, CD14+ cells, and CD15+ cells purified from the bone marrow of the same patients, using Affymetrix plus 2.0 microarrays. The five BMSC and seven osteoclast samples were obtained by culture in vitro. B. HPSE expression was measured by real-time RT-PCR. HPSE expression was normalized to that of GAPDH. One WBM sample was used as a reference and was assigned the arbitrary value of 100.
Figure 6. HPSE is expressed by BM environment cells and by a minor subpopulation of myeloma cells
A. XG-2 and XG-7 cells were stained with control rabbit polyclonal antibodies (Ab) (panel a) or with a polyclonal anti-HPSE Ab at 4.5 μg/ml (panels b,c). Note the dot-like staining signal. B. BM biopsies from patients with MM were stained with the polyclonal anti-HPSE Ab (4.5 μg/ml). Panels d–f: environment cells in the BM of one patient representative of twenty. PMN, polymorphonuclear cells; Meg, megakaryocytes. Panels g–j: heterogeneous HPSE expression among MMC in the bone marrow of four representative patients (g=patient 1; h=patient 4; i=patient 11; j=patient 20; see Table 3). The brown reaction product indicates the location of polyclonal Ab against HPSE. Original magnification of 1000× (or200× in f).
Figure 7. Event free survival and overall survival of patients with newly-diagnosed MM according to HPSE expression in the WBM and MMC
A. Expression of HPSE in the WBM of 30 newly diagnosed patients (left panel) and Kaplan-Meier survival curves for MM patients with the highest (_HPSE_high, n=15) or the lowest (_HPSE_low, n=15) HPSE expression in the WBM (right panel). B. Expression of HPSE in the purified MMC from 30 newly-diagnosed patients and Kaplan-Meier survival curves for MM patients with the highest (n=15) or the lowest (n=15) HPSE expression in the MMC.
Similar articles
- Heparanase enhances syndecan-1 shedding: a novel mechanism for stimulation of tumor growth and metastasis.
Yang Y, Macleod V, Miao HQ, Theus A, Zhan F, Shaughnessy JD Jr, Sawyer J, Li JP, Zcharia E, Vlodavsky I, Sanderson RD. Yang Y, et al. J Biol Chem. 2007 May 4;282(18):13326-33. doi: 10.1074/jbc.M611259200. Epub 2007 Mar 8. J Biol Chem. 2007. PMID: 17347152 - High heparanase activity in multiple myeloma is associated with elevated microvessel density.
Kelly T, Miao HQ, Yang Y, Navarro E, Kussie P, Huang Y, MacLeod V, Casciano J, Joseph L, Zhan F, Zangari M, Barlogie B, Shaughnessy J, Sanderson RD. Kelly T, et al. Cancer Res. 2003 Dec 15;63(24):8749-56. Cancer Res. 2003. PMID: 14695190 - The syndecan-1 heparan sulfate proteoglycan is a viable target for myeloma therapy.
Yang Y, MacLeod V, Dai Y, Khotskaya-Sample Y, Shriver Z, Venkataraman G, Sasisekharan R, Naggi A, Torri G, Casu B, Vlodavsky I, Suva LJ, Epstein J, Yaccoby S, Shaughnessy JD Jr, Barlogie B, Sanderson RD. Yang Y, et al. Blood. 2007 Sep 15;110(6):2041-8. doi: 10.1182/blood-2007-04-082495. Epub 2007 May 29. Blood. 2007. PMID: 17536013 Free PMC article. - Syndecan-1 and stromal heparan sulfate proteoglycans: key moderators of plasma cell biology and myeloma pathogenesis.
Ren Z, Spaargaren M, Pals ST. Ren Z, et al. Blood. 2021 Apr 1;137(13):1713-1718. doi: 10.1182/blood.2020008188. Blood. 2021. PMID: 33512430 Free PMC article. Review. - Role of Heparanase and Syndecan-1 in HSV-1 Release from Infected Cells.
Sharma P, Kapoor D, Shukla D. Sharma P, et al. Viruses. 2022 Sep 30;14(10):2156. doi: 10.3390/v14102156. Viruses. 2022. PMID: 36298711 Free PMC article. Review.
Cited by
- Relevance of serum levels of interleukin-6 and syndecan-1 in patients with hepatocellular carcinoma.
Metwaly HA, Al-Gayyar MM, Eletreby S, Ebrahim MA, El-Shishtawy MM. Metwaly HA, et al. Sci Pharm. 2012 Jan-Mar;80(1):179-88. doi: 10.3797/scipharm.1110-07. Epub 2011 Dec 23. Sci Pharm. 2012. PMID: 22396913 Free PMC article. - Heparanase: busy at the cell surface.
Fux L, Ilan N, Sanderson RD, Vlodavsky I. Fux L, et al. Trends Biochem Sci. 2009 Oct;34(10):511-9. doi: 10.1016/j.tibs.2009.06.005. Epub 2009 Sep 3. Trends Biochem Sci. 2009. PMID: 19733083 Free PMC article. Review. - Molecular functions of syndecan-1 in disease.
Teng YH, Aquino RS, Park PW. Teng YH, et al. Matrix Biol. 2012 Jan;31(1):3-16. doi: 10.1016/j.matbio.2011.10.001. Epub 2011 Oct 18. Matrix Biol. 2012. PMID: 22033227 Free PMC article. Review. - Heparan Sulfate Glycosaminoglycans: (Un)Expected Allies in Cancer Clinical Management.
Faria-Ramos I, Poças J, Marques C, Santos-Antunes J, Macedo G, Reis CA, Magalhães A. Faria-Ramos I, et al. Biomolecules. 2021 Jan 21;11(2):136. doi: 10.3390/biom11020136. Biomolecules. 2021. PMID: 33494442 Free PMC article. Review. - Heparanase inhibitor OGT 2115 induces prostate cancer cell apoptosis via the downregulation of MCL‑1.
Li X, Xu SJ, Jin B, Lu HS, Zhao SK, Ding XF, Xu LL, Li HJ, Liu SC, Chen J, Chen G. Li X, et al. Oncol Lett. 2024 Jan 5;27(2):83. doi: 10.3892/ol.2024.14217. eCollection 2024 Feb. Oncol Lett. 2024. PMID: 38249815 Free PMC article.
References
- Wijdenes J, Vooijs WC, Clement C, et al. A plasmocyte selective monoclonal antibody (B-B4) recognizes syndecan-1. Br J Haematol. 1996;94:318–323. - PubMed
- Costes V, Magen V, Legouffe E, et al. The Mi15 monoclonal antibody (anti-syndecan-1) is a reliable marker for quantifying plasma cells in paraffin-embedded bone marrow biopsy specimens. Hum Pathol. 1999;30:1405–1411. - PubMed
- Couchman JR. Syndecans: proteoglycan regulators of cell-surface microdomains? Nat Rev Mol Cell Biol. 2003;4:926–937. - PubMed
- Derksen PW, Keehnen RM, Evers LM, van Oers MH, Spaargaren M, Pals ST. Cell surface proteoglycan syndecan-1 mediates hepatocyte growth factor binding and promotes Met signaling in multiple myeloma. Blood. 2002;99:1405–1410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials