Alternative ion channel splicing in mesial temporal lobe epilepsy and Alzheimer's disease - PubMed (original) (raw)
Alternative ion channel splicing in mesial temporal lobe epilepsy and Alzheimer's disease
Erin L Heinzen et al. Genome Biol. 2007.
Abstract
Background: Alternative gene transcript splicing permits a single gene to produce multiple proteins with varied functions. Bioinformatic investigations have identified numerous splice variants, but whether these transcripts are translated to functional proteins and the physiological significance of these alternative proteins are largely unknown. Through direct identification of splice variants associated with disease states, we can begin to address these questions and to elucidate their roles in disease predisposition and pathophysiology. This work specifically sought to identify disease-associated alternative splicing patterns in ion channel genes by comprehensively screening affected brain tissue collected from patients with mesial temporal lobe epilepsy and Alzheimer's disease. New technology permitting the screening of alternative splice variants in microarray format was employed. Real time quantitative PCR was used to verify observed splice variant patterns.
Results: This work shows for the first time that two common neurological conditions are associated with extensive changes in gene splicing, with 25% and 12% of the genes considered having significant changes in splicing patterns associated with mesial temporal lobe epilepsy and Alzheimer's disease, respectively. Furthermore, these changes were found to exhibit unique and consistent patterns within the disease groups.
Conclusion: This work has identified a set of disease-associated, alternatively spliced gene products that represent high priorities for detailed functional investigations into how these changes impact the pathophysiology of mesial temporal lobe epilepsy and Alzheimer's disease.
Figures
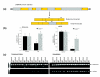
Figure 1
Representative mTLE-associated alternative splicing event identified using splice array technology. (a) Schematic of CLCN7 alternative splicing event associated with mTLE. Exons are shown in orange and intronic regions are shown in gray. (b) Data collected for the CLCN7 alternative splice event in control and mTLE brain tissue samples using the splice array technology (left) and quantitative rtPCR (qrtPCR, right). Data are presented as mean ± standard error of the mean; *p < 0.05 when compared to control, Student's _t_-test. (c) rtPCR confirmation of the pattern of transcript expression in brain tissue collected from ten subjects from each group. Both reference and variant transcript forms were amplified using the following primer sequences (indicated in the figure by arrows above mRNA transcripts): CLCN7F-GGCAAATACGCCCTGATG, CLCN7R-CTCAGCACGTCCACAATGAC.
Figure 2
Representative AD-associated alternative splicing event identified using splice array technology. (a) Schematic of GABRA6 alternative splicing event associated with mTLE. Exons are shown in orange and intronic regions are shown in gray. (b) Data collected for the GABRA6 alternative splice event in control and AD brain tissue (TC and CB) samples using the splice array technology (left) and quantitative rtPCR (qrtPCR, right). Data are presented as mean ± standard error of the mean; *p < 0.05 when compared to control, Student's _t_-test. (c) rtPCR confirmation of the pattern of transcript expression in brain tissue collected from ten subjects from each group. Reference and variant transcript forms were amplified using the following primer sequences (indicated in the figure by arrows above mRNA transcripts): reference GABRA6F-AAGAATCTTCAAGCCTTCTCCA, GABRA6R-TGACAGCTGCGAACTCGATA, variant GABRA6F-AAGAATCTTCAAGCCTTCTCCA, GABRA6F-TCCAAGATTACACAAATCTTTATATGC.
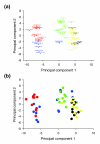
Figure 3
Principal component analysis of ion channel splice variant expression patterns for all experimental groups. (a) Colors separate groups based on statistical clustering (k-means clustering) of individuals with similar patterns of ion channel splicing. (b) Colors separate groups based on disease state and brain structure: control TC (blue), mTLE NC (red), AD TC (green), control CB (black), AD CB (yellow). The first principal component explains 43% of the variation in log expression ratios, while the second principal component accounts for 13% of the variation. Separation of clusters along principal components 1 and 2 is, to a large extent, governed by brain structure and disease state.
Similar articles
- Analysis of alternative splicing associated with aging and neurodegeneration in the human brain.
Tollervey JR, Wang Z, Hortobágyi T, Witten JT, Zarnack K, Kayikci M, Clark TA, Schweitzer AC, Rot G, Curk T, Zupan B, Rogelj B, Shaw CE, Ule J. Tollervey JR, et al. Genome Res. 2011 Oct;21(10):1572-82. doi: 10.1101/gr.122226.111. Epub 2011 Aug 16. Genome Res. 2011. PMID: 21846794 Free PMC article. - Upregulation of STREX splice variant of the large conductance Ca2+-activated potassium (BK) channel in a rat model of mesial temporal lobe epilepsy.
Ermolinsky BS, Skinner F, Garcia I, Arshadmansab MF, Otalora LF, Zarei MM, Garrido-Sanabria ER. Ermolinsky BS, et al. Neurosci Res. 2011 Jan;69(1):73-80. doi: 10.1016/j.neures.2010.09.011. Epub 2010 Oct 8. Neurosci Res. 2011. PMID: 20933547 Free PMC article. - Nova2 interacts with a cis-acting polymorphism to influence the proportions of drug-responsive splice variants of SCN1A.
Heinzen EL, Yoon W, Tate SK, Sen A, Wood NW, Sisodiya SM, Goldstein DB. Heinzen EL, et al. Am J Hum Genet. 2007 May;80(5):876-83. doi: 10.1086/516650. Epub 2007 Apr 3. Am J Hum Genet. 2007. PMID: 17436242 Free PMC article. - New Approaches to Studying Silent Mesial Temporal Lobe Seizures in Alzheimer's Disease.
Lam AD, Cole AJ, Cash SS. Lam AD, et al. Front Neurol. 2019 Sep 4;10:959. doi: 10.3389/fneur.2019.00959. eCollection 2019. Front Neurol. 2019. PMID: 31551916 Free PMC article. Review. - Potential Similarities in Temporal Lobe Epilepsy and Alzheimer’s Disease: From Clinic to Pathology.
Li BY, Chen SD. Li BY, et al. Am J Alzheimers Dis Other Demen. 2015 Dec;30(8):723-8. doi: 10.1177/1533317514537547. Am J Alzheimers Dis Other Demen. 2015. PMID: 24906967 Free PMC article. Review.
Cited by
- Revelation of Pivotal Genes Pertinent to Alzheimer's Pathogenesis: A Methodical Evaluation of 32 GEO Datasets.
Gns HS, Rajalekshmi SG, Burri RR. Gns HS, et al. J Mol Neurosci. 2022 Feb;72(2):303-322. doi: 10.1007/s12031-021-01919-2. Epub 2021 Oct 19. J Mol Neurosci. 2022. PMID: 34668150 Free PMC article. - Genome-wide association study of brain biochemical phenotypes reveals distinct genetic architecture of Alzheimer's disease related proteins.
Oatman SR, Reddy JS, Quicksall Z, Carrasquillo MM, Wang X, Liu CC, Yamazaki Y, Nguyen TT, Malphrus K, Heckman M, Biswas K, Nho K, Baker M, Martens YA, Zhao N, Kim JP, Risacher SL, Rademakers R, Saykin AJ, DeTure M, Murray ME, Kanekiyo T; Alzheimer’s Disease Neuroimaging Initiative; Dickson DW, Bu G, Allen M, Ertekin-Taner N. Oatman SR, et al. Mol Neurodegener. 2023 Jan 7;18(1):2. doi: 10.1186/s13024-022-00592-2. Mol Neurodegener. 2023. PMID: 36609403 Free PMC article. - Incorporating alternative splicing and mRNA editing into the genetic analysis of complex traits.
Hassan MA, Saeij JP. Hassan MA, et al. Bioessays. 2014 Nov;36(11):1032-40. doi: 10.1002/bies.201400079. Epub 2014 Aug 29. Bioessays. 2014. PMID: 25171292 Free PMC article. - A reliable and quick method for screening alternative splicing variants for low-abundance genes.
Zhang Y, Qu W, Yan R, Liu H, Zhang C, Li Z, Dong G. Zhang Y, et al. PLoS One. 2024 Jun 27;19(6):e0305201. doi: 10.1371/journal.pone.0305201. eCollection 2024. PLoS One. 2024. PMID: 38935635 Free PMC article. - Biophysical Kv3 channel alterations dampen excitability of cortical PV interneurons and contribute to network hyperexcitability in early Alzheimer's.
Olah VJ, Goettemoeller AM, Rayaprolu S, Dammer EB, Seyfried NT, Rangaraju S, Dimidschstein J, Rowan MJM. Olah VJ, et al. Elife. 2022 Jun 21;11:e75316. doi: 10.7554/eLife.75316. Elife. 2022. PMID: 35727131 Free PMC article.
References
- Glatz DC, Rujescu D, Tang Y, Berendt FJ, Hartmann AM, Faltraco F, Rosenberg C, Hulette C, Jellinger K, Hampel H. et al.The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and TRA2-BETA1 changes in sporadic Alzheimer's disease. J Neurochem. 2006;96:635–644. doi: 10.1111/j.1471-4159.2005.03552.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases