A functional study of miR-124 in the developing neural tube - PubMed (original) (raw)
A functional study of miR-124 in the developing neural tube
Xinwei Cao et al. Genes Dev. 2007.
Abstract
Neural development is a highly orchestrated process that entails precise control of gene expression. Although microRNAs (miRNAs) have been implicated in fine-tuning gene networks, the roles of individual miRNAs in vertebrate neural development have not been studied in vivo. We investigated the function of the most abundant neuronal miRNA, miR-124, during spinal cord development. Neither inhibition nor overexpression of miR-124 significantly altered the acquisition of neuronal fate, suggesting that miR-124 is unlikely to act as a primary determinant of neuronal differentiation. Two endogenous targets of miR-124, laminin gamma 1 and integrin beta1, were identified, both of which are highly expressed by neural progenitors but repressed upon neuronal differentiation. Thus miR-124 appears to ensure that progenitor genes are post-transcriptionally inhibited in neurons.
Figures
Figure 1.
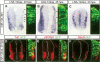
Inhibiting miR-124 does not affect neuronal differentiation. (A–C) miR-124 in situ hybridization (ISH) on chick embryos transfected with LNA-modified miR-124 antisense oligonucleotides (LNA-124as, 100 μM) showed decreased hybridization signals on the transfected side of the spinal cord at 20 and 45 hpe (arrows). Scrambled oligonucleotides (LNA-124scr) did not cause such a decrease. The image next to each ISH photograph shows the expression of the cotransfected GFP plasmid on an adjacent section. (D–F) Transfection of LNA-124as did not affect the expression of neuronal markers Tuj1, p27/Kip1, and NeuN at 20 hpe. Images on the left of the dashed lines show the whole neural tube sections, those on the right show only the transfected side.
Figure 2.
Overexpressing miR-124 in the chick neural tube. (A,B) The stem-loop structures of miR-124 and miR-124mt with red letters designating the sequences of mature miRNAs. Blue letters are the mutations introduced that should disrupt seed matches between miR-124 and its targets. (C) Northern blots probed for miR-124 or miR-124mt showed similar levels of expression from the overexpression plasmids in 293T cells. Lanes 1 and 4 are RNAs from untransfected 293T cells. Lanes 2, 3, 5, and 6 are RNAs from 293T cells transfected with the plasmid expressing miR-20, miR-124, miR-124, and miR-124mt, respectively. (D) miR-124 ISH showed high levels of expression (arrows) from the miR-124 plasmid in the transfected neural tube at 45 hpe. (E) The schematic drawing of a miRNA sensor plasmid. The two blue ovals represent two copies of complementary sequences to the miRNA that the sensor is designed to detect. The gray ovals represent scrambled sequences. (Pcag) Chick β-actin promoter; (d4EGFPn) nuclear-localized destabilized EGFP with a half-life of 4 h; (mRFPn) nuclear-localized monomeric red fluorescent protein. (F–H) At 45 hpe, most cells transfected with the sensor for miR-124 (pSensor-124) were yellow, expressing both GFP and RFP (F). Cotransfection of the miR-124 plasmid strongly repressed GFP expression (G), which was not observed with the miR-124mt plasmid (H).
Figure 3.
Overexpressing miR-124 does not promote overt neuronal differentiation. (A) Ectopic miR-124 did not affect the expression of the neural progenitor marker Sox2. (B) Cell cycle analysis by BrdU labeling. miR-124 was cotransfected with a plasmid expressing a nuclear localized GFP (GFPn) to facilitate cell counting. (C) Quantifications of the fraction of transfected cells (GFP+) that were mitotically active (BrdU+GFP+) showed no difference in embryos transfected with miR-124, miR-124mt, or the empty vector pENTR. (D–F) Overexpression of miR-124 did not increase the expression of neuronal markers Tuj1, p27/Kip1, or NeuN at 20 hpe.
Figure 4.
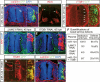
Ectopic miR-124 causes basal lamina defects. (A,B) Immunostaining for laminin-1 showed disintegration of basal laminae upon the overexpression of miR-124 (A, arrows). (B) The defect was located near the dorsal root entry site, shown on an adjacent section by immunostaining with BEN, an antibody that labels sensory axons (arrows). (C) The line of integrin β1 (ITGB1) that surrounds the outer surface of the neural tube was disrupted by ectopic miR-124 (arrows). (D,E) LAMC1 and ITGB1 RNAi caused similar, albeit milder, basal lamina defects (arrows). Transfected cells were marked by RFP encoded in RNAi vectors. (F) Quantifications of basal lamina defects. “Defects frequency” refers to the percentage of defective sections among all the sections examined in each embryo. (G,H) Cotransfection of an ITGB1-expressing plasmid with miR-124 partially rescued the basal lamina defect at 45 hpe (H). (G) High levels of ITGB1 expression were achieved from the transfected plasmid, with the overexpressed protein properly localized to the plasma membrane (inset).
Figure 5.
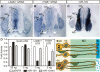
LAMC1 and ITGB1 mRNAs are endogenous targets of miR-124. (A–C) LAMC1, ITGB1, and miR-124 ISH on HH27–28 brachial-level transverse sections. (VZ) Ventricular zone; (MZ) mantle zone; (MD) motor domain; (DRG) dorsal root ganglion. (D) Luciferase assays with the luciferase gene fused to SV40, chick LAMC1, or chick ITGB1 3′-UTR. pENTR, miR-124, or miR-124mt plasmids (amounts shown in the _X_-axis) were cotransfected with luciferase–UTR constructs. Luciferase activities obtained from the cotransfection of pENTR and each luciferase–UTR construct were used as denominators. Cotransfection of miR-124, but not miR-124mt, specifically repressed LAMC1 and ITGB1 UTRs but not SV40 UTR. (E, panel a) During neural tube development, neural progenitors (orange) contribute to the formation and maintenance of basal laminae (BL) by producing laminin γ1 (black balls) and integrin β1 (red bars). (Panel b) Upon ectopic expression of miR-124 (short purple lines) in neural progenitors, both genes were repressed, leading to the disintegration of basal laminae. Green cells represent differentiating neurons.
Similar articles
- MicroRNA-mediated control of oligodendrocyte differentiation.
Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, Wang F, Appel B, Lu QR. Zhao X, et al. Neuron. 2010 Mar 11;65(5):612-26. doi: 10.1016/j.neuron.2010.02.018. Neuron. 2010. PMID: 20223198 Free PMC article. - Regulation of miRNA expression during neural cell specification.
Smirnova L, Gräfe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Smirnova L, et al. Eur J Neurosci. 2005 Mar;21(6):1469-77. doi: 10.1111/j.1460-9568.2005.03978.x. Eur J Neurosci. 2005. PMID: 15845075 - The on/off of Pax6 controls the tempo of neuronal differentiation in the developing spinal cord.
Bel-Vialar S, Medevielle F, Pituello F. Bel-Vialar S, et al. Dev Biol. 2007 May 15;305(2):659-73. doi: 10.1016/j.ydbio.2007.02.012. Epub 2007 Feb 16. Dev Biol. 2007. PMID: 17399698 - Transcriptional networks regulating neuronal identity in the developing spinal cord.
Lee SK, Pfaff SL. Lee SK, et al. Nat Neurosci. 2001 Nov;4 Suppl:1183-91. doi: 10.1038/nn750. Nat Neurosci. 2001. PMID: 11687828 Review. - Establishing neuronal diversity in the spinal cord: a time and a place.
Sagner A, Briscoe J. Sagner A, et al. Development. 2019 Nov 25;146(22):dev182154. doi: 10.1242/dev.182154. Development. 2019. PMID: 31767567 Review.
Cited by
- Bta-miR-181d and Bta-miR-196a mediated proliferation, differentiation, and apoptosis in Bovine Myogenic Cells.
Chengcheng L, Raza SHA, Zhimei Y, Sihu W, Shengchen Y, Aloufi BH, Bingzhi L, Zan L. Chengcheng L, et al. J Anim Sci. 2024 Jan 3;102:skae142. doi: 10.1093/jas/skae142. J Anim Sci. 2024. PMID: 38766769 - Communication between the stem cell niche and an adjacent differentiation niche through miRNA and EGFR signaling orchestrates exit from the stem cell state in the Drosophila ovary.
Chen J, Li C, Sheng Y, Zhang J, Pang L, Dong Z, Wu Z, Lu Y, Liu Z, Zhang Q, Guan X, Chen X, Huang J. Chen J, et al. PLoS Biol. 2024 Mar 21;22(3):e3002515. doi: 10.1371/journal.pbio.3002515. eCollection 2024 Mar. PLoS Biol. 2024. PMID: 38512963 Free PMC article. - G9a inactivation in progenitor cells with Isl1-Cre with reduced recombinase activity models aspects of Dandy-Walker complex.
Chi L, Zhong L, Lee D, Yu X, Caballero A, Nieman B, Delgado-Olguin P. Chi L, et al. Biol Open. 2023 Aug 15;12(8):bio059894. doi: 10.1242/bio.059894. Epub 2023 Jul 28. Biol Open. 2023. PMID: 37470706 Free PMC article. - MicroRNA‑124: an emerging therapeutic target in central nervous system disorders.
Zhang WH, Jiang L, Li M, Liu J. Zhang WH, et al. Exp Brain Res. 2023 May;241(5):1215-1226. doi: 10.1007/s00221-022-06524-2. Epub 2023 Mar 24. Exp Brain Res. 2023. PMID: 36961552 Free PMC article. Review. - Modeling Movement Disorders via Generation of hiPSC-Derived Motor Neurons.
Akter M, Ding B. Akter M, et al. Cells. 2022 Nov 27;11(23):3796. doi: 10.3390/cells11233796. Cells. 2022. PMID: 36497056 Free PMC article. Review.
References
- Bartel D.P., Chen C.Z., Chen C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. - PubMed
- Bylund M., Andersson E., Novitch B.G., Muhr J., Andersson E., Novitch B.G., Muhr J., Novitch B.G., Muhr J., Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 2003;6:1162–1168. - PubMed
- Chen C.Z., Li L., Lodish H.F., Bartel D.P., Li L., Lodish H.F., Bartel D.P., Lodish H.F., Bartel D.P., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. - PubMed
- Das R.M., Van Hateren N.J., Howell G.R., Farrell E.R., Bangs F.K., Porteous V.C., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., Van Hateren N.J., Howell G.R., Farrell E.R., Bangs F.K., Porteous V.C., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., Howell G.R., Farrell E.R., Bangs F.K., Porteous V.C., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., Farrell E.R., Bangs F.K., Porteous V.C., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., Bangs F.K., Porteous V.C., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., Porteous V.C., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., McGrew M.J., Ohyama K., Sacco M.A., Ohyama K., Sacco M.A., Sacco M.A. A robust system for RNA interference in the chicken using a modified microRNA operon. Dev. Biol. 2006;294:554–563. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous