Evolution and the complexity of bacteriophages - PubMed (original) (raw)
Evolution and the complexity of bacteriophages
Philip Serwer. Virol J. 2007.
Abstract
Background: The genomes of both long-genome (> 200 Kb) bacteriophages and long-genome eukaryotic viruses have cellular gene homologs whose selective advantage is not explained. These homologs add genomic and possibly biochemical complexity. Understanding their significance requires a definition of complexity that is more biochemically oriented than past empirically based definitions.
Hypothesis: Initially, I propose two biochemistry-oriented definitions of complexity: either decreased randomness or increased encoded information that does not serve immediate needs. Then, I make the assumption that these two definitions are equivalent. This assumption and recent data lead to the following four-part hypothesis that explains the presence of cellular gene homologs in long bacteriophage genomes and also provides a pathway for complexity increases in prokaryotic cells: (1) Prokaryotes underwent evolutionary increases in biochemical complexity after the eukaryote/prokaryote splits. (2) Some of the complexity increases occurred via multi-step, weak selection that was both protected from strong selection and accelerated by embedding evolving cellular genes in the genomes of bacteriophages and, presumably, also archaeal viruses (first tier selection). (3) The mechanisms for retaining cellular genes in viral genomes evolved under additional, longer-term selection that was stronger (second tier selection). (4) The second tier selection was based on increased access by prokaryotic cells to improved biochemical systems. This access was achieved when DNA transfer moved to prokaryotic cells both the more evolved genes and their more competitive and complex biochemical systems.
Testing the hypothesis: I propose testing this hypothesis by controlled evolution in microbial communities to (1) determine the effects of deleting individual cellular gene homologs on the growth and evolution of long genome bacteriophages and hosts, (2) find the environmental conditions that select for the presence of cellular gene homologs, (3) determine which, if any, bacteriophage genes were selected for maintaining the homologs and (4) determine the dynamics of homolog evolution.
Implications of the hypothesis: This hypothesis is an explanation of evolutionary leaps in general. If accurate, it will assist both understanding and influencing the evolution of microbes and their communities. Analysis of evolutionary complexity increase for at least prokaryotes should include analysis of genomes of long-genome bacteriophages.
Figures
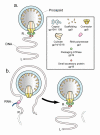
Figure 1
Initiation of DNA packaging by the closely related bacteriophages, T3 and T7. (a) Initiation is illustrated for the simplest DNA packaging. This packaging has a monomeric DNA substrate and was demonstrated for T3 and assumed for T7. Packaging of this type occurs only in vitro, as far as is known (review [31,32]). (b) Initiation is illustrated for the more complex DNA packaging that occurs in vivo for both T3 and T7 (review [31,32]). In (b), the DNA substrate is an end-to-end joined concatemeric DNA for which only one monomer is completely shown. Dashed lines in (b) indicate part of another monomer within the concatemer. The following details of the concatemer are omitted for simplicity: replication forks and interaction among different procapsids (review [32]). The various proteins and protein assemblies of the initiation complex, including the connector and DNA packaging ATPase, are identified in the rectangular box. Proteins have both descriptive names and names based on gene number [2], preceded by gp. The letter, R, indicates the right end of the mature DNA molecule; the letter, L, indicates the left end.
Similar articles
- Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life.
Prangishvili D, Garrett RA, Koonin EV. Prangishvili D, et al. Virus Res. 2006 Apr;117(1):52-67. doi: 10.1016/j.virusres.2006.01.007. Epub 2006 Feb 28. Virus Res. 2006. PMID: 16503363 Review. - Identification and investigation of ORFans in the viral world.
Yin Y, Fischer D. Yin Y, et al. BMC Genomics. 2008 Jan 19;9:24. doi: 10.1186/1471-2164-9-24. BMC Genomics. 2008. PMID: 18205946 Free PMC article. - Complete nucleotide sequence and likely recombinatorial origin of bacteriophage T3.
Pajunen MI, Elizondo MR, Skurnik M, Kieleczawa J, Molineux IJ. Pajunen MI, et al. J Mol Biol. 2002 Jun 21;319(5):1115-32. doi: 10.1016/S0022-2836(02)00384-4. J Mol Biol. 2002. PMID: 12079351 - The genome sequence of Yersinia pestis bacteriophage phiA1122 reveals an intimate history with the coliphage T3 and T7 genomes.
Garcia E, Elliott JM, Ramanculov E, Chain PS, Chu MC, Molineux IJ. Garcia E, et al. J Bacteriol. 2003 Sep;185(17):5248-62. doi: 10.1128/JB.185.17.5248-5262.2003. J Bacteriol. 2003. PMID: 12923098 Free PMC article. - Evolutionary genomics of nucleo-cytoplasmic large DNA viruses.
Iyer LM, Balaji S, Koonin EV, Aravind L. Iyer LM, et al. Virus Res. 2006 Apr;117(1):156-84. doi: 10.1016/j.virusres.2006.01.009. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16494962 Review.
Cited by
- The discovery of phiAGATE, a novel phage infecting Bacillus pumilus, leads to new insights into the phylogeny of the subfamily Spounavirinae.
Barylski J, Nowicki G, Goździcka-Józefiak A. Barylski J, et al. PLoS One. 2014 Jan 23;9(1):e86632. doi: 10.1371/journal.pone.0086632. eCollection 2014. PLoS One. 2014. PMID: 24466180 Free PMC article. - DNA packaging motor assembly intermediate of bacteriophage phi29.
Koti JS, Morais MC, Rajagopal R, Owen BA, McMurray CT, Anderson DL. Koti JS, et al. J Mol Biol. 2008 Sep 19;381(5):1114-32. doi: 10.1016/j.jmb.2008.04.034. Epub 2008 Apr 20. J Mol Biol. 2008. PMID: 18674782 Free PMC article. - Classification of Myoviridae bacteriophages using protein sequence similarity.
Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann HW, Kropinski AM. Lavigne R, et al. BMC Microbiol. 2009 Oct 26;9:224. doi: 10.1186/1471-2180-9-224. BMC Microbiol. 2009. PMID: 19857251 Free PMC article. - In-Gel Isolation and Characterization of Large (and Other) Phages.
Serwer P, Wright ET. Serwer P, et al. Viruses. 2020 Apr 7;12(4):410. doi: 10.3390/v12040410. Viruses. 2020. PMID: 32272774 Free PMC article. Review. - Dualities in the analysis of phage DNA packaging motors.
Serwer P, Jiang W. Serwer P, et al. Bacteriophage. 2012 Oct 1;2(4):239-255. doi: 10.4161/bact.23829. Bacteriophage. 2012. PMID: 23532204 Free PMC article.
References
- Jardine PJ, Anderson DL. DNA packaging in dsDNA bacteriophages. In: Calendar R, editor. The Bacteriophages. New York: Oxford University Press; 2006.
- Kutter E, Stidham T, Guttman B, Kutter E, Batts D, Peterson S, Djavakhishvili T, Arisaka F, Mesyanzhinov V, Rüger W, Mosig G. Genomic map of bacteriophage T4. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. Washington, DC: ASM Press; 1994. pp. 491–519.
- Miller ES, Heidelberg JF, Eisen JA, Nelson WC, Durkin AS, Ciecko A, Feldblyum TV, White O, Paulsen IT, Nierman WC, Lee J, Szczypinski B, Fraser CM. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J Bacteriol. 2003;185:5220–5233. doi: 10.1128/JB.185.17.5220-5233.2003. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous