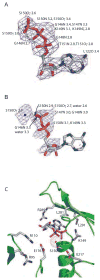Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility - PubMed (original) (raw)
Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility
Kenneth A Satyshur et al. Structure. 2007 Mar.
Abstract
PilT is a hexameric ATPase required for bacterial type IV pilus retraction and surface motility. Crystal structures of ADP- and ATP-bound Aquifex aeolicus PilT at 2.8 and 3.2 A resolution show N-terminal PAS-like and C-terminal RecA-like ATPase domains followed by a set of short C-terminal helices. The hexamer is formed by extensive polar subunit interactions between the ATPase core of one monomer and the N-terminal domain of the next. An additional structure captures a nonsymmetric PilT hexamer in which approach of invariant arginines from two subunits to the bound nucleotide forms an enzymatically competent active site. A panel of pilT mutations highlights the importance of the arginines, the PAS-like domain, the polar subunit interface, and the C-terminal helices for retraction. We present a model for ATP binding leading to dramatic PilT domain motions, engagement of the arginine wire, and subunit communication in this hexameric motor. Our conclusions apply to the entire type II/IV secretion ATPase family.
Figures
Figure 1
The sequence and secondary structure of A. aeolicus PilT (AaPilT) aligned with P. aeruginosa PilT (PaPilT), N. gonorrhoeae PilT (NgPilT), V. cholerae EpsE (EpsE) and H. pylori HP0525 (HP0525). Alignments are based on 3-D superpositions, sequence homology, and manual optimization. Observed A. aeolicus PilT α-helices (h) and β-strands (>) are denoted above the sequence, and indicated by blue and red letters, respectively, for the three known structures. Type II/IV secretion ATPase motifs are indicated by shaded boxes: Walker A, Asp Box, Walker B and His Box, in that order. Other symbols mark the CTDn:NTDn+1 interface (diamond), recognition of adenine (plus sign), arginine wire (quotation), other residues presumed to participate in ATP recognition and catalysis (pound sign), and positions of changes in other non-functional PilT variants from this study (dagger). Dots indicate every 10th amino acid in the A. aeolicus PilT sequence. The EpsE zinc-binding tetracysteine motif (CM subdomain) is marked by a carat; the alignment continues after this 40 amino acid insertion. Residues not resolved in the refined structures are typed in grey. EpsE and HP0525 sequences are not shown upstream of the region of structural homology.
Figure 2
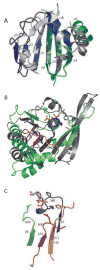
Structure of PilT monomer. (A) Stereo view of the refined structure of the PilT:ADP monomer. The N-terminal PAS-like domain is blue and cyan to indicate quasi internal 2-fold symmetry. The CTD secondary structure elements form the RecA fold (green), helices αH and αI which form the hexamer constriction and are an insertion in the RecA fold (magenta), helices αJ and αK (red), and the far C-terminal helices αL, αM, and αN (grey). Conserved residues Arg110, Lys149, Ser 150 and Glu176, as well as the bound water and ADP, signpost the nucleotide-binding pocket. (B) Schematic of the PilT secondary structure elements with conserved ATP binding motifs indicated. The coloring scheme is as in (A).
Figure 3
Conserved elements of PilT fold. (A) The NTD of PilT (colored as in Fig. 2A) resembles the well-known PAS domain (grey, represented by the circadian clock protein Period (Yildiz et al., 2005)). Non-canonical PAS elements (PilT αA and loops within Period) are removed for clarity. (B) The core ATPase subdomain of PilT (green) is readily superimposable upon RecA (Story and Steitz, 1992) (grey). In this view the least squares calculation is over P-loop residues only. Type II/IV secretion ATPase family motifs Walker A (blue), Asp Box (lime green), Walker B (magenta) and His Box (orange) neighbor the bound nucleotide. (C) Isolated, magnified view of the four sequence motifs described in (B), with ADP and signature invariant residues Lys149, Glu176, Glu217 and His242 depicted (blue, green, magenta, and orange, respectively).
Figure 4
Nucleotide binding in PilT. Composite Fo-Fc omit electron density maps show (A) ATP (countoured at 2.0σ) and (B) ADP (contoured at 1.0σ) in the nucleotide-binding site of the respective PilT P6 structures. Protein atoms within 3.4 Å of ATP are enumerated in (A). In (B), the water is also indicated and protein atoms within 3.4 Å of the water, phosphates, or ribose are enumerated. (C) Close-up view of side chains and water adjacent to the nucleotide binding site in PilT:ADP, with backbone ribbon colored as in Figure 1A.
Figure 5
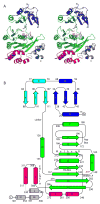
Hexamer formation. (A) The P6 space group constrains the hexamer to be symmetric, as shown in this view of the assembly in which one subunit is highlighted in yellow. (B) In both ADP and ATP-bound P6 crystal forms, the main intermolecular interface is CTDn:NTDn+1 as seen in these two adjacent subunits, viewed perpendicular to the 6-fold symmetry axis. Black outlines the non-covalent CTDn:NTDn+1 pair. (C) Zoomed-in stereo view of the intermolecular CTDn:NTDn+1 interface, rotated to highlight a subset of the amino acids in the polar interface. The coloring in all panels is as in Figure 2.
Figure 6
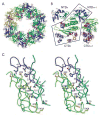
Dramatic domain orientation differences in the PilT C2 hexamer. (A) 6-fold symmetry is broken. Subunits E and B (light and dark blue, respectively) are displaced from the center of the hexamer. (B) Adjacent subunits E (blue) and F (orange) were superimposed over CTD residues 116–361. The NTDs of these monomers are then related by a 69° rotation about an axis through the domain linker. Helices C are labeled to highlight the exent of the movement. (C) Close-up view of the nucleotide-binding site at the three-way interface of NTDF (orange, below) and CTDF (orange, above) from the closed, liganded subunit and the CTDE (blue) from the adjacent open subunit, with refined ADP and modeled Pi shown in Fo-Fc omit electron density calculated without nucleotide (contoured at 2.5σ (blue) and 5σ (red)). Note the arginine wire. Given the low resolution of the data used for this refinement, it is appropriate to consider this figure as one model that is consistent with our data. (D) At the CTDE:CTDF interface, arginine finger residues are engaged and 950 Å2 are buried.
Figure 7
Protein levels of variant PilTs in vivo. Western blot analysis, using a polyclonal anti-P. aeruginosa PilT antibody, was used to assess relative levels of plasmid-encoded PilT in a PAK pilT background. The binding of the antibody to an unidentified constant band at higher molecular weight serves as a loading control. The wild type protein is shown with full and half-strength sample concentrations (WT and WT 1/2). Samples come from P. aeruginosa cells expressing the indicated PilT protein.
Figure 8
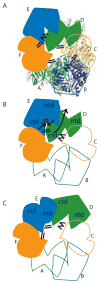
Model for concerted PilT motions. (A) The quasi 2-fold symmetric C2 crystal structure has two peripheral wide-open subunits (B, E; blue), two central “active” subunits (C, F; orange) and two central “resting” subunits (A, D; green). Four CTD:CTD interfaces have engaged arginine fingers (double lines). The remaining two have disengaged fingers (zig-zag). Subunit F is clamped around bound nucleotide. (B) When ATP (red) binds in the E cleft, the two domains close around the ligand (short black arrows), causing the β5/β6 arginines to approach the ATP. Because of the extensive CTDD:NTDE interface, the motion of NTDE forces the swiveling of CTDD (in particular the C-terminal helices) toward the periphery of the hexamer (long grey arrow). Consequently, the D arginine fingers engage in the E active site (double lines). On the other side of CTDD, the CTDC:CTDD interface likewise rearranges, disengaging the C arginine fingers from the D active site (zig-zag). (C) Subunit D is now poised as the most peripheral, wide open subunit and ready to bind nucleotide; E is clamped around nucleotide and contributing to an engaged CTD:CTD interface on either side.
Comment in
- Secretion superfamily ATPases swing big.
Savvides SN. Savvides SN. Structure. 2007 Mar;15(3):255-7. doi: 10.1016/j.str.2007.02.003. Structure. 2007. PMID: 17355860 No abstract available.
Similar articles
- Functional dissection of a conserved motif within the pilus retraction protein PilT.
Aukema KG, Kron EM, Herdendorf TJ, Forest KT. Aukema KG, et al. J Bacteriol. 2005 Jan;187(2):611-8. doi: 10.1128/JB.187.2.611-618.2005. J Bacteriol. 2005. PMID: 15629932 Free PMC article. - Structural insights into the mechanism of Type IVa pilus extension and retraction ATPase motors.
Solanki V, Kapoor S, Thakur KG. Solanki V, et al. FEBS J. 2018 Sep;285(18):3402-3421. doi: 10.1111/febs.14619. Epub 2018 Aug 18. FEBS J. 2018. PMID: 30066435 - Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU.
Chiang P, Sampaleanu LM, Ayers M, Pahuta M, Howell PL, Burrows LL. Chiang P, et al. Microbiology (Reading). 2008 Jan;154(Pt 1):114-126. doi: 10.1099/mic.0.2007/011320-0. Microbiology (Reading). 2008. PMID: 18174131 - The PIN-domain toxin-antitoxin array in mycobacteria.
Arcus VL, Rainey PB, Turner SJ. Arcus VL, et al. Trends Microbiol. 2005 Aug;13(8):360-5. doi: 10.1016/j.tim.2005.06.008. Trends Microbiol. 2005. PMID: 15993073 Review. - Pulling together with type IV pili.
Nudleman E, Kaiser D. Nudleman E, et al. J Mol Microbiol Biotechnol. 2004;7(1-2):52-62. doi: 10.1159/000077869. J Mol Microbiol Biotechnol. 2004. PMID: 15170403 Review.
Cited by
- Systematic functional analysis reveals that a set of seven genes is involved in fine-tuning of the multiple functions mediated by type IV pili in Neisseria meningitidis.
Brown DR, Helaine S, Carbonnelle E, Pelicic V. Brown DR, et al. Infect Immun. 2010 Jul;78(7):3053-63. doi: 10.1128/IAI.00099-10. Epub 2010 May 3. Infect Immun. 2010. PMID: 20439474 Free PMC article. - Electron Cryotomography of Bacterial Secretion Systems.
Oikonomou CM, Jensen GJ. Oikonomou CM, et al. Microbiol Spectr. 2019 Mar;7(2):10.1128/microbiolspec.psib-0019-2018. doi: 10.1128/microbiolspec.PSIB-0019-2018. Microbiol Spectr. 2019. PMID: 30953431 Free PMC article. - Oligomerization of EpsE coordinates residues from multiple subunits to facilitate ATPase activity.
Patrick M, Korotkov KV, Hol WG, Sandkvist M. Patrick M, et al. J Biol Chem. 2011 Mar 25;286(12):10378-86. doi: 10.1074/jbc.M110.167031. Epub 2011 Jan 5. J Biol Chem. 2011. PMID: 21209100 Free PMC article. - Combinatorial control of type IVa pili formation by the four polarized regulators MglA, SgmX, FrzS, and SopA.
Oklitschek M, Carreira LAM, Muratoğlu M, Søgaard-Andersen L, Treuner-Lange A. Oklitschek M, et al. J Bacteriol. 2024 Nov 21;206(11):e0010824. doi: 10.1128/jb.00108-24. Epub 2024 Oct 15. J Bacteriol. 2024. PMID: 39404445 Free PMC article. - Zinc coordination is essential for the function and activity of the type II secretion ATPase EpsE.
Rule CS, Patrick M, Camberg JL, Maricic N, Hol WG, Sandkvist M. Rule CS, et al. Microbiologyopen. 2016 Oct;5(5):870-882. doi: 10.1002/mbo3.376. Epub 2016 May 10. Microbiologyopen. 2016. PMID: 27168165 Free PMC article.
References
- Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 Å resolution of the F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. - PubMed
- Arcus VL, Bäckbro|| K, Roos|| A, Daniel EL, Baker EN. Distant Structural Homology Leads to the Functional Characterization of an Archaeal PIN Domain as an Exonuclease. J Biol Chem. 2004:16471–16478. - PubMed
- Brunger A, Adams P, Clore M, Gros P, Nilges M, Read R. Crystallography & NMR System. Acta Cryst. 1998;D54:905–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases