Proteomic methods for analysis of S-nitrosation - PubMed (original) (raw)
Review
Proteomic methods for analysis of S-nitrosation
Nicholas J Kettenhofen et al. J Chromatogr B Analyt Technol Biomed Life Sci. 2007.
Abstract
This review discusses proteomic methods to detect and identify S-nitrosated proteins. Protein S-nitrosation, the post-translational modification of thiol residues to form S-nitrosothiols, has been suggested to be a mechanism of cellular redox signaling by which nitric oxide can alter cellular function through modification of protein thiol residues. It has become apparent that methods that will detect and identify low levels of S-nitrosated protein in complex protein mixtures are required in order to fully appreciate the range, extent and selectivity of this modification in both physiological and pathological conditions. While many advances have been made in the detection of either total cellular S-nitrosation or individual S-nitrosothiols, proteomic methods for the detection of S-nitrosation are in relative infancy. This review will discuss the major methods that have been used for the proteomic analysis of protein S-nitrosation and discuss the pros and cons of this methodology.
Figures
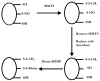
Fig. 1
The biotin switch assay (after Jaffrey et al. [42]). Protein thiols (-SH), _S_-nitrosothiols (-SNO) and disulfides (-SSR) are treated with MMTS to convert all thiols to disulfides (-SSCH3). MMTS is removed and the proteins are incubated with ascorbate to convert –SNO to –SH. The nascent –SH is then labeled with biotin via a disulfide bond using biotin-HPDP.
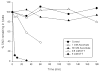
Fig. 2
Reduction of protein RSNO by ascorbate and DTT. Human bronchial epithelial cells were incubated with CysNO (50 μM) for 30 minutes. Cells were lyzed in HEPES buffer containing 10 mM NEM, 100 μM DTPA and 10 μM neocuproine. Either DTT or ascorbate was added, and aliquots were taken at the indicated time points. For each aliquot, the protein was precipitated to remove excess reducing agent, and total RSNO was measured by tri-iodide-based chemiluminescence after treatment with sulfanilamide.
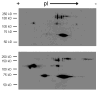
Fig. 3
Biotinylation of proteins after DTT reduction. Human bronchial epithelial cells were incubated without (top) or with (bottom) 10 μM CysNO and then subjected to the biotin switch assay using DTT (5 mM) in place of ascorbate. Proteins were separated in 2 dimensions and biotinylation was examined by Western blot analysis.
Fig. 4
Analysis of protein modification by DIGE. (Top) Schematic of the DIGE labeling scheme using two maleimido-cyanine-based dyes Cy3 (green) and Cy5 (red). RSNO or NO treatment will modify some fraction of the protein thiols as illustrated on the left side of the figure. Both the treated and untreated samples are sequentially incubated with NEM, reducing agent (in this case DTT) and then either Cy3 or Cy5. These samples are pooled at equal protein concentrations are run in 2 dimensions. (Bottom) Human bronchial epithelial cells were treated with or without 5 μM CysNO, and protein was treated as illustrated in the top panel. Pooled protein was run in 2 dimensions and detected by fluorescence scanning.
Similar articles
- Detection and proteomic identification of S-nitrosated proteins in human hepatocytes.
López-Sánchez LM, Corrales FJ, De La Mata M, Muntané J, Rodríguez-Ariza A. López-Sánchez LM, et al. Methods Enzymol. 2008;440:273-81. doi: 10.1016/S0076-6879(07)00817-8. Methods Enzymol. 2008. PMID: 18423224 Review. - Characterization and application of the biotin-switch assay for the identification of S-nitrosated proteins.
Zhang Y, Keszler A, Broniowska KA, Hogg N. Zhang Y, et al. Free Radic Biol Med. 2005 Apr 1;38(7):874-81. doi: 10.1016/j.freeradbiomed.2004.12.012. Free Radic Biol Med. 2005. PMID: 15749383 - Proteomic and mass spectroscopic quantitation of protein S-nitrosation differentiates NO-donors.
Sinha V, Wijewickrama GT, Chandrasena RE, Xu H, Edirisinghe PD, Schiefer IT, Thatcher GR. Sinha V, et al. ACS Chem Biol. 2010 Jul 16;5(7):667-80. doi: 10.1021/cb100054m. ACS Chem Biol. 2010. PMID: 20524644 Free PMC article. - Proteomic approaches to evaluate protein S-nitrosylation in disease.
López-Sánchez LM, López-Pedrera C, Rodríguez-Ariza A. López-Sánchez LM, et al. Mass Spectrom Rev. 2014 Jan-Feb;33(1):7-20. doi: 10.1002/mas.21373. Epub 2013 Jun 15. Mass Spectrom Rev. 2014. PMID: 23775552 Review.
Cited by
- Redox signaling and protein phosphorylation in mitochondria: progress and prospects.
Foster DB, Van Eyk JE, Marbán E, O'Rourke B. Foster DB, et al. J Bioenerg Biomembr. 2009 Apr;41(2):159-68. doi: 10.1007/s10863-009-9217-7. J Bioenerg Biomembr. 2009. PMID: 19440831 Free PMC article. Review. - Detection of protein S-nitrosylation with the biotin-switch technique.
Forrester MT, Foster MW, Benhar M, Stamler JS. Forrester MT, et al. Free Radic Biol Med. 2009 Jan 15;46(2):119-26. doi: 10.1016/j.freeradbiomed.2008.09.034. Epub 2008 Oct 17. Free Radic Biol Med. 2009. PMID: 18977293 Free PMC article. Review. - Quantitative site-specific reactivity profiling of S-nitrosylation in mouse skeletal muscle using cysteinyl peptide enrichment coupled with mass spectrometry.
Su D, Shukla AK, Chen B, Kim JS, Nakayasu E, Qu Y, Aryal U, Weitz K, Clauss TR, Monroe ME, Camp DG 2nd, Bigelow DJ, Smith RD, Kulkarni RN, Qian WJ. Su D, et al. Free Radic Biol Med. 2013 Apr;57:68-78. doi: 10.1016/j.freeradbiomed.2012.12.010. Epub 2012 Dec 28. Free Radic Biol Med. 2013. PMID: 23277143 Free PMC article. - A reductive ligation based fluorescent probe for S-nitrosothiols.
Zhang D, Chen W, Miao Z, Ye Y, Zhao Y, King SB, Xian M. Zhang D, et al. Chem Commun (Camb). 2014 May 14;50(37):4806-9. doi: 10.1039/c4cc01288g. Chem Commun (Camb). 2014. PMID: 24658175 Free PMC article. - Nitric oxide-based protein modification: formation and site-specificity of protein S-nitrosylation.
Kovacs I, Lindermayr C. Kovacs I, et al. Front Plant Sci. 2013 May 14;4:137. doi: 10.3389/fpls.2013.00137. eCollection 2013. Front Plant Sci. 2013. PMID: 23717319 Free PMC article.
References
- Stamler JS, Lamas S, Fang FC. Cell. 2001;106:675. - PubMed
- Hogg N, Singh RJ, Kalyanaraman B. FEBS Lett. 1996;382:223. - PubMed
- Folkes LK, Wardman P. Free Radic Biol Med. 2004;37:549. - PubMed
- DeMaster EG, Quast BJ, Redfern B, Nagasawa HT. Biochemistry. 1995;34:11494. - PubMed
- Pryor WA, Church DF, Govindan CK, Crank G. J Org Chem. 1982;47:159.
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM055792-09/GM/NIGMS NIH HHS/United States
- R01 GM055792/GM/NIGMS NIH HHS/United States
- R01 GM055792-10A2/GM/NIGMS NIH HHS/United States
- R29 GM055792/GM/NIGMS NIH HHS/United States
- GM55792/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous