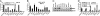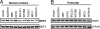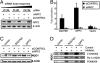A genomic screen for activators of the antioxidant response element - PubMed (original) (raw)
A genomic screen for activators of the antioxidant response element
Yanxia Liu et al. Proc Natl Acad Sci U S A. 2007.
Abstract
The antioxidant response element (ARE) is a cis-acting regulatory enhancer element found in the 5' flanking region of many phase II detoxification enzymes. Up-regulation of ARE-dependent target genes is known to have neuroprotective effects; yet, the mechanism of activation is largely unknown. By screening an arrayed collection of approximately 15,000 full-length expression cDNAs in the human neuroblastoma cell line IMR-32 with an ARE-luciferase reporter, we have identified several cDNAs not previously associated with ARE activation. A subset of cDNAs, encoding sequestosome 1 (SQSTM1) and dipeptidylpeptidase 3 (DPP3), activated the ARE in primary mouse-derived cortical neurons. Overexpression of SQSTM1 and DPP3 in IMR-32 cells stimulated NF-E2-related factor 2 (NRF2) nuclear translocation and led to increased levels of NAD(P)H:quinone oxidoreductase 1, a protein which is transcriptionally regulated by the ARE. When transfected into IMR-32 neuroblastoma cells that were depleted of transcription factor NRF2 by RNA interference, SQSTM1 and DPP3 were unable to activate the ARE or induce NAD(P)H:quinone oxidoreductase 1 expression, indicating that the ARE activation upon ectopic expression of these cDNAs is mediated by NRF2. Studies with pharmacological inhibitors indicated that 1-phosphatidylinositol 3-kinase and protein kinase C signaling are essential for activity. Overexpression of these cDNAs conferred partial resistance to hydrogen peroxide or rotenone-induced toxicity, consistent with the induction of antioxidant and phase II detoxification enzymes, which can protect from oxidative stress. This work and other such studies may provide mechanisms for activating the ARE in the absence of general oxidative stress and a yet-unexploited therapeutic approach to degenerative diseases and aging.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
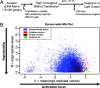
Genome-wide cDNA overexpression screen for ARE activators. (A) General high-throughput screening procedure. Approximately 15,000 expression cDNAs, normalized and arrayed in 384-well plates, were transfected into IMR-32 human neuroblastoma cells along with an ARE–luciferase reporter construct. After 48-h incubation, luciferase activity was assessed by measuring luminescence output per well. (B) Screen-wide MA plot. The screen was carried out in duplicate, and M (a measure of screen-to-screen variation; σ = standard deviation) was plotted as a function of A (a measure of the mean ARE activation from both screens). The cDNAs that strongly activated the ARE in a reproducible manner were investigated further (indicated in green, lower right corner).
Fig. 2.
Confirmation of putative screening hits in IMR-32 cells and mouse primary cortical culture. (A) Transcriptional ARE activation in IMR-32 cells with wild-type and mutant enhancer elements. IMR-32 cells were cotransfected with cDNAs, the ARE-luciferase reporter (wild-type or mutant), and actin-lacZ for normalization in 24-well plates. Luminescence was detected 48 h later; normalized values are given (n = 6). Eight cDNAs showed equal or higher activity than the positive control, PI3K*. (B) Effect of cDNA overexpression in IMR-32 cells on NQO1 transcript levels as analyzed by quantitative real-time PCR (qPCR). The cDNAs were introduced by lipofection, and then total RNA was isolated 48 h later, reverse-transcribed to cDNA, and subjected to TaqMan analysis (n = 3). GAPDH expression was used as internal control for normalization. Overexpression of several cDNAs increased NQO1 levels to the same extent as Nrf2 overexpression. (C) Induction of NQO1 upon cDNA overexpression in IMR-32 cells as analyzed by Western blot analysis. cDNAs were transfected by using lipofection, and proteins were isolated 48 h later, resolved by SDS/PAGE, and subjected to Western blot analysis for NQO1. SQSTM1 and DPP3 induced NQO1 most strongly and to a comparable extent as Nrf2. A representative blot (n = 4) is shown. (D) Transcriptional ARE activation in mouse primary cortical culture. After 5 days, cells were transfected with the reporter mixture (ARE–luciferase, CMV-lacZ, and cDNA), and 24 h later cells were treated either with 10 μM tBHQ or with vehicle control. After an additional 24 h, luminescence was detected. Normalized values are given (n = 3). Only a subset of cDNAs with activity in IMR-32 cells showed activity in primary cells. The ARE activity of SQSTM1 and DPP3 could not be further augmented by tBHQ.
Fig. 3.
Analysis of NRF2 translocation upon cDNA overexpression in IMR-32 cells. (A) Enrichment of NRF2 in the nucleus by SQSTM1 and DPP3 overexpression as determined by immunoblot analysis. Nuclear extracts prepared with the NE-PER reagent kit (Pierce) 48 h after transfection were resolved by SDS/PAGE, and the Western blot was probed with NRF2 antibody. A representative blot (n = 3) is shown. SQSTM1 and DPP3 significantly increased NRF2 content in the nucleus. Levels of the nuclear protein OCT1 did not change. (B) Effect of cDNA overexpression on NRF2 transcript levels. Total RNA from IMR-32 cells was harvested 48 h after cDNA transfection. Semiquantitative RT-PCR suggested that NRF2 transcript levels did not change significantly. Data shown are representative of multiple experiments (n = 3) in the linear amplification range.
Fig. 4.
Effect of siRNAs targeting NRF2 and effects of kinase inhibitors in IMR-32 cells. (A) Dose-response analysis for siRNAs by using RT-PCR. SiLentFect-mediated transfection of 50 nM siRNAs almost completely abrogated NRF2 transcript levels after 48 h in a reproducible manner (n = 4). (B) Effect of SQSTM1 and DPP3 overexpression in NRF2-depleted IMR-32 cells. The cDNAs encoding SQSTM1 and DPP3 (1.4 μg) were cotransfected with ARE-luciferase reporter (1.0 μg) and with siRNAs against NRF2 or control siRNAs (50 nM) in six-well plate format. After 48 h, luciferase activity was measured. DNA transfection efficiency was monitored by cotransfecting CMV–GFP (0.2 μg) and was similar in each well. SQSTM1 and DPP3 were unable to activate the ARE in cells transfected with siRNAs against NRF2 (n = 4). Note that ARE activation by SQSTM1 and DPP3 in the control cells is lower than that shown in Fig. 2_A_ because less cDNA was transfected in this experiment, consistent with a dose-dependency of ARE activation. (C) Analysis of SQSTM1 and DPP3 mediated induction of NQO1 in NRF2-depleted IMR-32 cells. The cDNAs encoding SQSTM1 and DPP3 (1.4 μg) were cotransfected with siRNAs against NRF2 or control siRNAs (50 nM) in six-well plate format. Equal DNA transfection efficiency was obtained by monitoring GFP expression upon cotransfection with CMV–GFP (0.2 μg). SQSTM1 and DPP3 were unable to induce NQO1 expression in cells cotransfected with siRNAs targeting the NRF2 transcript. Nontargeting siRNAs did not diminish the ability of SQSTM1 and DPP3 to induce NQO1. Control protein levels (β-actin) did not change across experimental conditions. A representative blot (n = 3) is shown. (D) Effect of pharmacological kinase inhibitors on SQSTM1- and DPP3-induced NQO1 expression. IMR-32 cells were seeded in six-well plates (600,000 cells per well) and 1 day later were transfected with cDNA (1.6 μg) and CMV–GFP (0.4 μg) by using siLentFect. After 24 h, cells were treated for additional 24 h with PI3K inhibitor LY294002 (25 μM), PKC inhibitor Ro-31-8220 (1 μM), and MEK1 inhibitor PD98059 (50 μM). Concentrations used have been shown to be effective (14, 17). Representative blots (n = 3) are shown.
Fig. 5.
Nrf2, SQSTM1 and DPP3 mediate protection from oxidative stress in vitro. In 24-well plate format with siLentFect, 180,000 IMR-32 cells were transfected with corresponding cDNAs and with 0.5 μg of vector control. After 48 h, cells were treated with various concentrations of hydrogen peroxide for 6 h (A) or rotenone for 12 h (B) (n = 4). Cell viability was assessed by using the CellTiter-Glo assay kit (Promega). Overexpression of Nrf2, SQSTM1, or DPP3 attenuated the toxic effects of both stressors. ∗ indicates statistical significance compared with vector (P < 0.05).
Similar articles
- Nrf2-dependent activation of the antioxidant responsive element by tert-butylhydroquinone is independent of oxidative stress in IMR-32 human neuroblastoma cells.
Lee JM, Moehlenkamp JD, Hanson JM, Johnson JA. Lee JM, et al. Biochem Biophys Res Commun. 2001 Jan 12;280(1):286-92. doi: 10.1006/bbrc.2000.4106. Biochem Biophys Res Commun. 2001. PMID: 11162512 - Shear stress stabilizes NF-E2-related factor 2 and induces antioxidant genes in endothelial cells: role of reactive oxygen/nitrogen species.
Warabi E, Takabe W, Minami T, Inoue K, Itoh K, Yamamoto M, Ishii T, Kodama T, Noguchi N. Warabi E, et al. Free Radic Biol Med. 2007 Jan 15;42(2):260-9. doi: 10.1016/j.freeradbiomed.2006.10.043. Epub 2006 Oct 20. Free Radic Biol Med. 2007. PMID: 17189831 - Nrf2 signaling in coordinated activation of antioxidant gene expression.
Jaiswal AK. Jaiswal AK. Free Radic Biol Med. 2004 May 15;36(10):1199-207. doi: 10.1016/j.freeradbiomed.2004.02.074. Free Radic Biol Med. 2004. PMID: 15110384 Review. - Regulatory mechanisms controlling gene expression mediated by the antioxidant response element.
Nguyen T, Sherratt PJ, Pickett CB. Nguyen T, et al. Annu Rev Pharmacol Toxicol. 2003;43:233-60. doi: 10.1146/annurev.pharmtox.43.100901.140229. Epub 2002 Jan 10. Annu Rev Pharmacol Toxicol. 2003. PMID: 12359864 Review.
Cited by
- DPP3 expression promotes cell proliferation and migration in vitro and tumour growth in vivo, which is associated with poor prognosis of oesophageal carcinoma.
Liu JK, Abudula A, Yang HT, Xu LX, Nuerrula Y, Bai G, Tulahong A, Eli M. Liu JK, et al. Oncol Rep. 2023 Jan;49(1):9. doi: 10.3892/or.2022.8446. Epub 2022 Nov 16. Oncol Rep. 2023. PMID: 36382663 Free PMC article. - Oxidative stress response and Nrf2 signaling in aging.
Zhang H, Davies KJA, Forman HJ. Zhang H, et al. Free Radic Biol Med. 2015 Nov;88(Pt B):314-336. doi: 10.1016/j.freeradbiomed.2015.05.036. Epub 2015 Jun 9. Free Radic Biol Med. 2015. PMID: 26066302 Free PMC article. Review. - Transcriptional analysis of apoptotic cerebellar granule neurons following rescue by gastric inhibitory polypeptide.
Maino B, Ciotti MT, Calissano P, Cavallaro S. Maino B, et al. Int J Mol Sci. 2014 Apr 1;15(4):5596-622. doi: 10.3390/ijms15045596. Int J Mol Sci. 2014. PMID: 24694544 Free PMC article. - The effect of 17β-estradiol on the expression of dipeptidyl peptidase III and heme oxygenase 1 in liver of CBA/H mice.
Mačak Šafranko Ž, Sobočanec S, Šarić A, Jajčanin-Jozić N, Krsnik Ž, Aralica G, Balog T, Abramić M. Mačak Šafranko Ž, et al. J Endocrinol Invest. 2015 Apr;38(4):471-9. doi: 10.1007/s40618-014-0217-z. Epub 2014 Nov 29. J Endocrinol Invest. 2015. PMID: 25432329 - Quantitative proteomics reveals a "poised quiescence" cellular state after triggering the DNA replication origin activation checkpoint.
Mulvey C, Tudzarova S, Crawford M, Williams GH, Stoeber K, Godovac-Zimmermann J. Mulvey C, et al. J Proteome Res. 2010 Oct 1;9(10):5445-60. doi: 10.1021/pr100678k. J Proteome Res. 2010. PMID: 20707412 Free PMC article.
References
- van Muiswinkel FL, Kuiperij HB. Curr Drug Targets CNS Neurol Disord. 2005;4:267–281. - PubMed
- Nguyen T, Sherratt PJ, Pickett CB. Annu Rev Pharmacol Toxicol. 2003;43:233–260. - PubMed
- Li J, Lee JM, Johnson JA. J Biol Chem. 2002;277:388–394. - PubMed
- Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. J Biol Chem. 2003;278:12029–12038. - PubMed
- Johnson DA, Andrews GK, Xu W, Johnson JA. J Neurochem. 2002;81:1233–1241. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 ES008089/ES/NIEHS NIH HHS/United States
- R29 ES008089/ES/NIEHS NIH HHS/United States
- R01 ES010042/ES/NIEHS NIH HHS/United States
- ES10042/ES/NIEHS NIH HHS/United States
- ES08089/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous