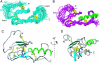Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand - PubMed (original) (raw)
Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand
Christy Rani R Grace et al. Proc Natl Acad Sci U S A. 2007.
Abstract
The corticotropin releasing factor (CRF) family of ligands and their receptors coordinate endocrine, behavioral, autonomic, and metabolic responses to stress and play additional roles within the cardiovascular, gastrointestinal, and other systems. The actions of CRF and the related urocortins are mediated by activation of two receptors, CRF-R1 and CRF-R2, belonging to the B1 family of G protein-coupled receptors. The short-consensus-repeat fold (SCR) within the first extracellular domain (ECD1) of the CRF receptor(s) comprises the major ligand binding site and serves to dock a peptide ligand via its C-terminal segment, thus positioning the N-terminal segment to interact with the receptor's juxtamembrane domains to activate the receptor. Here we present the 3D NMR structure of ECD1 of CRF-R2beta in complex with astressin, a peptide antagonist. In the structure of the complex the C-terminal segment of astressin forms an amphipathic helix, whose entire hydrophobic face interacts with the short-consensus-repeat motif, covering a large intermolecular interface. In addition, the complex is characterized by intermolecular hydrogen bonds and a salt bridge. These interactions are quantitatively weighted by an analysis of the effects on the full-length receptor affinities using an Ala scan of CRF. These structural studies identify the major determinants for CRF ligand specificity and selectivity and support a two-step model for receptor activation. Furthermore, because of a proposed conservation of the fold for both the ECD1s and ligands, this structure can serve as a model for ligand recognition for the entire B1 receptor family.
Conflict of interest statement
Conflict of interest statement: W.W.V. is a cofounder, consultant, equity holder, and member of the Board of Directors of Neurocrine Biosciences and Acceleron Pharma. The following have been licensed by The Salk Institute for Biological Studies and/or The Clayton Foundation: CRF to Ferring Pharmaceuticals, CRF1 receptor and Ucn 2 to Neurocrine Biosciences, and Ucn 3 to Johnson & Johnson.
Figures
Fig. 1.
3D NMR structure of ECD1–CRF-R2β in complex with astressin. (A) Superposition of 20 energy-minimized conformers representing the 3D NMR structure of free ECD1–CRF-R2β (the backbone Cα atoms of residues 57–83 and 99–120 were superimposed). (B) Superposition of 20 energy-minimized conformers of ECD1–CRF-R2β in complex with astressin (the backbone Cα atoms of residues 57–120 of ECD1 and 30–41 of astressin were superimposed). The backbone of residues 44–122 of ECD1 is shown in magenta, and the backbone of residues Leu-27–Ile-41 of astressin is colored in green. In A and B the disulfide bonds are shown in yellow. (C) Ribbon diagram of the lowest energy conformer representing the 3D NMR structure of the ECD–CRF-R2β–astressin complex. The β-sheets are shown in cyan, and the side chains of the core residues Trp-71 and Trp-109 along with the disulfide bonds are shown in yellow. The salt bridge Arg-101 (in blue)–Asp-65 (in red) is shown as dashed spine in orange. The backbone of astressin from Leu-27–Ile-41 is shown in green. (D) Side view of the ribbon diagram shown in C. MOLMOL was used to generate the figures (49).
Fig. 2.
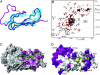
Structural and dynamical characterization of the ECD–CRF-R2β–astressin complex. (A) Superposition of the 20 conformers of free ECD1–CRF-R2β depicted as a cyan sausage with the lowest energy conformer of the complex ECD1–CRF-R2β–astressin shown as a magenta ribbon. The disulfide bonds are shown in yellow. (B) [15N,1H]-TROSY spectra of the free (in red contours) and complex (in black contours) ECD1–CRF-R2β. Residues enclosed in rectangular boxes are from loop 2 (residues 84–98). The peaks from free ECD1–CRF-R2β are missing for most of the residues in loop 2 because of slow dynamics. (C) Surface representation of ECD1–CRF-R2β showing the residues in pink, whose chemical shift perturbations are >0.5 ppm upon complex formation with astressin (see also
SI Fig. 4_C_
). Astressin is shown in green along with the side chains interacting with the ECD1. (D) Amino acid sequence differences of mouse CRF-R1 and CRF-R2 are mapped in purple onto the ECD1 surface of the complex structure. The residues are labeled with single letter code for CRF-R1, followed by CRF-R2. Astressin is shown as a green–yellow ribbon. Highlighted in yellow are the sequence differences of CRF ligands (see also
SI Fig. 6
).
Fig. 3.
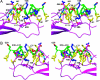
Molecular anatomy of the residues in the interaction site between astressin and ECD1–CRF-R2β. Shown are front (A) and back (B) stereo views of the side chain interactions between the ECD1 and astressin. The backbones of astressin and ECD1 are shown as green and magenta ribbons, respectively. Hydrophobic side chains are shown in yellow, hydrogen bonds are shown in cyan, and the salt bridges Arg-35–Glu-86 and Lys-36–Glu-39 are shown in gray. All of the residues in the interaction surface are marked for clarity. “X” refers to norleucine residue (Nle).
Similar articles
- The three-dimensional structure of the N-terminal domain of corticotropin-releasing factor receptors: sushi domains and the B1 family of G protein-coupled receptors.
Perrin MH, Grace CR, Riek R, Vale WW. Perrin MH, et al. Ann N Y Acad Sci. 2006 Jul;1070:105-19. doi: 10.1196/annals.1317.065. Ann N Y Acad Sci. 2006. PMID: 16888152 - Ligand affinity for amino-terminal and juxtamembrane domains of the corticotropin releasing factor type I receptor: regulation by G-protein and nonpeptide antagonists.
Hoare SR, Sullivan SK, Schwarz DA, Ling N, Vale WW, Crowe PD, Grigoriadis DE. Hoare SR, et al. Biochemistry. 2004 Apr 6;43(13):3996-4011. doi: 10.1021/bi036110a. Biochemistry. 2004. PMID: 15049707 - NMR structure of the first extracellular domain of corticotropin-releasing factor receptor 1 (ECD1-CRF-R1) complexed with a high affinity agonist.
Grace CR, Perrin MH, Gulyas J, Rivier JE, Vale WW, Riek R. Grace CR, et al. J Biol Chem. 2010 Dec 3;285(49):38580-9. doi: 10.1074/jbc.M110.121897. Epub 2010 Sep 15. J Biol Chem. 2010. PMID: 20843795 Free PMC article. - Corticotropin releasing factor receptors and their ligand family.
Perrin MH, Vale WW. Perrin MH, et al. Ann N Y Acad Sci. 1999 Oct 20;885:312-28. doi: 10.1111/j.1749-6632.1999.tb08687.x. Ann N Y Acad Sci. 1999. PMID: 10816663 Review. - Members of CRF family and their receptors: from past to future.
Liapakis G, Venihaki M, Margioris A, Grigoriadis D, Gkountelias K. Liapakis G, et al. Curr Med Chem. 2011;18(17):2583-600. doi: 10.2174/092986711795933704. Curr Med Chem. 2011. PMID: 21568890 Review.
Cited by
- Mutational and cysteine scanning analysis of the glucagon receptor N-terminal domain.
Prévost M, Vertongen P, Raussens V, Roberts DJ, Cnudde J, Perret J, Waelbroeck M. Prévost M, et al. J Biol Chem. 2010 Oct 1;285(40):30951-8. doi: 10.1074/jbc.M110.102814. Epub 2010 Jul 20. J Biol Chem. 2010. PMID: 20647307 Free PMC article. - Molecular basis of glucagon-like peptide 1 docking to its intact receptor studied with carboxyl-terminal photolabile probes.
Chen Q, Pinon DI, Miller LJ, Dong M. Chen Q, et al. J Biol Chem. 2009 Dec 4;284(49):34135-44. doi: 10.1074/jbc.M109.038109. Epub 2009 Oct 8. J Biol Chem. 2009. PMID: 19815559 Free PMC article. - Effects of pH and temperature on photoaffinity labeling of Family B G protein-coupled receptors.
Dong M, Miller LJ. Dong M, et al. Regul Pept. 2009 Nov 27;158(1-3):110-5. doi: 10.1016/j.regpep.2009.05.007. Epub 2009 May 18. Regul Pept. 2009. PMID: 19454296 Free PMC article. - Spatial approximations between residues 6 and 12 in the amino-terminal region of glucagon-like peptide 1 and its receptor: a region critical for biological activity.
Chen Q, Pinon DI, Miller LJ, Dong M. Chen Q, et al. J Biol Chem. 2010 Aug 6;285(32):24508-18. doi: 10.1074/jbc.M110.135749. Epub 2010 Jun 7. J Biol Chem. 2010. PMID: 20529866 Free PMC article. - Residue 17 of sauvagine cross-links to the first transmembrane domain of corticotropin-releasing factor receptor 1 (CRFR1).
Assil-Kishawi I, Samra TA, Mierke DF, Abou-Samra AB. Assil-Kishawi I, et al. J Biol Chem. 2008 Dec 19;283(51):35644-51. doi: 10.1074/jbc.M806351200. Epub 2008 Oct 27. J Biol Chem. 2008. PMID: 18955489 Free PMC article.
References
- Vale W, Spiess J, Rivier C, Rivier J. Science. 1981;213:1394–1397. - PubMed
- Armario A. CNS Neurol Disord Drug Targets. 2006;5:485–501. - PubMed
- Madden J, Newton S. Clin J Oncol Nurs. 2006;10:659–661. - PubMed
- Bale TL, Contarino A, Smith GW, Chan R, Gold LH, Sawchenko PE, Koob GF, Vale WW, Lee K-F. Nat Gen. 2000;24:410–414. - PubMed
- Smith GW, Aubry J-M, Dellu F, Contarino A, Bilezikjian LM, Gold L, Chen R, Marchuk Y, Hauser C, Bentley CA, et al. Neuron. 1998;20:1093–1102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases