Accurate, conformation-dependent predictions of solvent effects on protein ionization constants - PubMed (original) (raw)
Accurate, conformation-dependent predictions of solvent effects on protein ionization constants
P Barth et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Predicting how aqueous solvent modulates the conformational transitions and influences the pKa values that regulate the biological functions of biomolecules remains an unsolved challenge. To address this problem, we developed FDPB_MF, a rotamer repacking method that exhaustively samples side chain conformational space and rigorously calculates multibody protein-solvent interactions. FDPB_MF predicts the effects on pKa values of various solvent exposures, large ionic strength variations, strong energetic couplings, structural reorganizations and sequence mutations. The method achieves high accuracy, with root mean square deviations within 0.3 pH unit of the experimental values measured for turkey ovomucoid third domain, hen lysozyme, Bacillus circulans xylanase, and human and Escherichia coli thioredoxins. FDPB_MF provides a faithful, quantitative assessment of electrostatic interactions in biological macromolecules.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
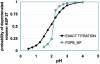
Accuracy of the convergence of the FDPB_MF compared with the solution obtained by exhaustively sampling all of the discrete states of the system. The two different predicted titration curves of Asp 27 correspond to the sum of the probability of the deprotonated rotamers of Asp-27 as a function of the pH. The “exact titration” curve (black) was obtained by enumerating all of the discrete states of the system and is equal to the sum at a given pH of the Boltzmann weights of the discrete states occupied by the deprotonated rotamers. The FDPB_MF curve (gray) is derived by relaxing a rotameric probability distribution with the FDPB_MF method. It corresponds to the probability that minimizes at a given pH the electrostatic free energy of the protein after optimization by the SCMF algorithm.
Fig. 2.
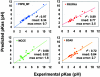
Synopsis of the pKa predictions performed by FDPB_MF and comparison with other methods: PROPKA (18), MCCE (10), and EGAD (9). rsmds and maximal errors to the experimental values are in pH units. Each line and _r_2 value corresponds to the best linear regression fit to the data and the correlation coefficient, respectively.
Fig. 3.
Good agreement between observed and predicted conformational changes induced by the protonation of Glu 172 in B. circulans xylanase. Figures were generated by using Pymol (
). As discussed in ref. , the crystal structure solved at an apparent pH of 4.0 is likely to be consistent with Glu 172 being protonated. Protons on the x-ray structures were added with the program Reduce (32). At the bottom, distance changes are provided in angstroms. The first and second numbers represent distances between protons and heteroatoms and distances between heteroatoms, respectively.
Similar articles
- Toward accurate prediction of pKa values for internal protein residues: the importance of conformational relaxation and desolvation energy.
Wallace JA, Wang Y, Shi C, Pastoor KJ, Nguyen BL, Xia K, Shen JK. Wallace JA, et al. Proteins. 2011 Dec;79(12):3364-73. doi: 10.1002/prot.23080. Epub 2011 Jul 11. Proteins. 2011. PMID: 21748801 - Tanford-Kirkwood electrostatics for protein modeling.
Havranek JJ, Harbury PB. Havranek JJ, et al. Proc Natl Acad Sci U S A. 1999 Sep 28;96(20):11145-50. doi: 10.1073/pnas.96.20.11145. Proc Natl Acad Sci U S A. 1999. PMID: 10500144 Free PMC article. - Protein side-chain rearrangement in regions of point mutations.
Eyal E, Najmanovich R, Edelman M, Sobolev V. Eyal E, et al. Proteins. 2003 Feb 1;50(2):272-82. doi: 10.1002/prot.10276. Proteins. 2003. PMID: 12486721 - Electrostatics calculations: latest methodological advances.
Koehl P. Koehl P. Curr Opin Struct Biol. 2006 Apr;16(2):142-51. doi: 10.1016/j.sbi.2006.03.001. Epub 2006 Mar 15. Curr Opin Struct Biol. 2006. PMID: 16540310 Review. - Structural basis of perturbed pKa values of catalytic groups in enzyme active sites.
Harris TK, Turner GJ. Harris TK, et al. IUBMB Life. 2002 Feb;53(2):85-98. doi: 10.1080/15216540211468. IUBMB Life. 2002. PMID: 12049200 Review.
Cited by
- A physics-based energy function allows the computational redesign of a PDZ domain.
Opuu V, Sun YJ, Hou T, Panel N, Fuentes EJ, Simonson T. Opuu V, et al. Sci Rep. 2020 Jul 7;10(1):11150. doi: 10.1038/s41598-020-67972-w. Sci Rep. 2020. PMID: 32636412 Free PMC article. - Toward high-resolution prediction and design of transmembrane helical protein structures.
Barth P, Schonbrun J, Baker D. Barth P, et al. Proc Natl Acad Sci U S A. 2007 Oct 2;104(40):15682-7. doi: 10.1073/pnas.0702515104. Epub 2007 Sep 28. Proc Natl Acad Sci U S A. 2007. PMID: 17905872 Free PMC article. - Designing specific protein-protein interactions using computation, experimental library screening, or integrated methods.
Chen TS, Keating AE. Chen TS, et al. Protein Sci. 2012 Jul;21(7):949-63. doi: 10.1002/pro.2096. Epub 2012 Jun 8. Protein Sci. 2012. PMID: 22593041 Free PMC article. Review. - Using polarizable POSSIM force field and fuzzy-border continuum solvent model to calculate pK(a) shifts of protein residues.
Sharma I, Kaminski GA. Sharma I, et al. J Comput Chem. 2017 Jan 15;38(2):65-80. doi: 10.1002/jcc.24519. Epub 2016 Oct 27. J Comput Chem. 2017. PMID: 27785788 Free PMC article. - Factors affecting the use of 13C(alpha) chemical shifts to determine, refine, and validate protein structures.
Vila JA, Scheraga HA. Vila JA, et al. Proteins. 2008 May 1;71(2):641-54. doi: 10.1002/prot.21726. Proteins. 2008. PMID: 17975838 Free PMC article.
References
- Garcia-Moreno BE, Fitch CA. Methods Enzymol. 2004;380:20–51. - PubMed
- Schutz CN, Warshel A. Proteins. 2001;44:400–417. - PubMed
- Baker NA. Methods Enzymol. 2004;383:94–118. - PubMed
- Baker NA. Curr Opin Struct Biol. 2005;15:137–143. - PubMed
- Mongan J, Case DA. Curr Opin Struct Biol. 2005;15:157–163. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources