Targeted access to the genomes of low-abundance organisms in complex microbial communities - PubMed (original) (raw)
Targeted access to the genomes of low-abundance organisms in complex microbial communities
Mircea Podar et al. Appl Environ Microbiol. 2007 May.
Abstract
Current metagenomic approaches to the study of complex microbial consortia provide a glimpse into the community metabolism and occasionally allow genomic assemblies for the most abundant organisms. However, little information is gained for the members of the community present at low frequencies, especially those representing yet-uncultured taxa, which include the bulk of the diversity present in most environments. Here we used phylogenetically directed cell separation by fluorescence in situ hybridization and flow cytometry, followed by amplification and sequencing of a fraction of the genomic DNA of several bacterial cells that belong to the TM7 phylum. Partial genomic assembly allowed, for the first time, a look into the evolution and potential metabolism of a soil representative from this group of organisms for which there are no species in stable laboratory cultures. Genomic reconstruction from targeted cells of uncultured organisms isolated directly from the environment represents a powerful approach to access any specific members of a community and an alternative way to assess the community's metabolic potential.
Figures
FIG. 1.
(A) Taxonomic distribution (domain/phylum level) of SSU rRNA sequences in the PCR clone library prepared from the soil sample. (B) Flow cytometric analysis of soil bacteria hybridized with a fluorescent TM7-specific oligonucleotide probe (TM7905) or in the absence of hybridization (Control). The inner rectangle indicates the gating used for separation, and the arrow points to the low-frequency, highly fluorescent cells in the hybridized sample that were separated.
FIG. 2.
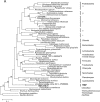
(A) Phylogenetic analysis (maximum likelihood) showing the relationships between the SSU rRNA sequence of TM7_GTL1 and previously known environmental clone sequences representing the three TM7 subdomains (27), with sequences from TG1 and Chloroflexi as outgroups. Filled circles represent bootstrap support values over 50% (shown only for major nodes). The bar indicates inferred 10% sequence divergence. GenBank accession numbers are given. (B) Secondary-structure model of TM7_GTL1 SSU rRNA, with the P37_2 helix shaded.
FIG. 3.
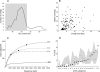
Information content analyses for the TM7_GTL1 genomic library. (A) Average GC content (percent) distribution of the assembled primary contigs. The shaded region indicates the contigs that were pooled as representing the TM7_GTL1 genome. Contigs with GC contents of >53% represent a Pseudomonas minor coisolate. (B) Average depth coverages (_n_-fold) for the final TM7 contigs. (C) Functional accumulation curves (COG categories) for TM7_GTL1 and representative finished bacterial genomes as a function of sequence depth (sequencing reads). The filled circle indicates the sequence depth corresponding to a 1× coverage for each of the finished genomes. P.u., Pelagibacter ubique; G.s., Geobacter sulfurreducens; C.t., Chlorobium tepidum. (D) Percentages of distribution of major COG categories in the TM7_GTL1 data (filled circles). For comparison, the median frequency values based on 502 bacterial genomes (not including obligatory parasites and symbionts) are indicated by the open circles. The gray area spans the observed distribution of frequencies in those genomes, between the minimum and maximum values.
FIG. 4.
Phylogenetic analysis (maximum likelihood) of the relationships among bacterial phyla, determined using SSU rRNA (A) and concatenated ribosomal protein sequences (B). Bootstrap support values over 50 are indicated (for major nodes only). The placement of TM7_GTL1 is indicated by shaded labels. rRNA sequence 1277396 represents the Pseudomonas coisolate of the amplified TM7_GTL1 genomic DNA. The bar indicates inferred 10% sequence divergence.
FIG. 4.
Phylogenetic analysis (maximum likelihood) of the relationships among bacterial phyla, determined using SSU rRNA (A) and concatenated ribosomal protein sequences (B). Bootstrap support values over 50 are indicated (for major nodes only). The placement of TM7_GTL1 is indicated by shaded labels. rRNA sequence 1277396 represents the Pseudomonas coisolate of the amplified TM7_GTL1 genomic DNA. The bar indicates inferred 10% sequence divergence.
Similar articles
- A novel retrieval system for nearly complete microbial genomic fragments from soil samples.
Yu WH, Su SC, Lee CY. Yu WH, et al. J Microbiol Methods. 2008 Feb;72(2):197-205. doi: 10.1016/j.mimet.2007.11.022. Epub 2007 Dec 8. J Microbiol Methods. 2008. PMID: 18187218 - Phylogenetic and metagenomic analysis of Verrucomicrobia in former agricultural grassland soil.
Kielak A, Rodrigues JL, Kuramae EE, Chain PS, van Veen JA, Kowalchuk GA. Kielak A, et al. FEMS Microbiol Ecol. 2010 Jan;71(1):23-33. doi: 10.1111/j.1574-6941.2009.00785.x. FEMS Microbiol Ecol. 2010. PMID: 19811538 - Cultivating the uncultured.
Zengler K, Toledo G, Rappe M, Elkins J, Mathur EJ, Short JM, Keller M. Zengler K, et al. Proc Natl Acad Sci U S A. 2002 Nov 26;99(24):15681-6. doi: 10.1073/pnas.252630999. Epub 2002 Nov 18. Proc Natl Acad Sci U S A. 2002. PMID: 12438682 Free PMC article. - The uncultured microbial majority.
Rappé MS, Giovannoni SJ. Rappé MS, et al. Annu Rev Microbiol. 2003;57:369-94. doi: 10.1146/annurev.micro.57.030502.090759. Annu Rev Microbiol. 2003. PMID: 14527284 Review. - Microbial diversity in the era of omic technologies.
Nikolaki S, Tsiamis G. Nikolaki S, et al. Biomed Res Int. 2013;2013:958719. doi: 10.1155/2013/958719. Epub 2013 Oct 24. Biomed Res Int. 2013. PMID: 24260747 Free PMC article. Review.
Cited by
- Artificial polyploidy improves bacterial single cell genome recovery.
Dichosa AE, Fitzsimons MS, Lo CC, Weston LL, Preteska LG, Snook JP, Zhang X, Gu W, McMurry K, Green LD, Chain PS, Detter JC, Han CS. Dichosa AE, et al. PLoS One. 2012;7(5):e37387. doi: 10.1371/journal.pone.0037387. Epub 2012 May 22. PLoS One. 2012. PMID: 22666352 Free PMC article. - Building on basic metagenomics with complementary technologies.
Warnecke F, Hugenholtz P. Warnecke F, et al. Genome Biol. 2007;8(12):231. doi: 10.1186/gb-2007-8-12-231. Genome Biol. 2007. PMID: 18177506 Free PMC article. Review. - How sulphate-reducing microorganisms cope with stress: lessons from systems biology.
Zhou J, He Q, Hemme CL, Mukhopadhyay A, Hillesland K, Zhou A, He Z, Van Nostrand JD, Hazen TC, Stahl DA, Wall JD, Arkin AP. Zhou J, et al. Nat Rev Microbiol. 2011 Jun;9(6):452-66. doi: 10.1038/nrmicro2575. Epub 2011 May 16. Nat Rev Microbiol. 2011. PMID: 21572460 Review. - Bioprospecting Sponge-Associated Microbes for Antimicrobial Compounds.
Indraningrat AA, Smidt H, Sipkema D. Indraningrat AA, et al. Mar Drugs. 2016 May 2;14(5):87. doi: 10.3390/md14050087. Mar Drugs. 2016. PMID: 27144573 Free PMC article. Review. - Novelty and uniqueness patterns of rare members of the soil biosphere.
Elshahed MS, Youssef NH, Spain AM, Sheik C, Najar FZ, Sukharnikov LO, Roe BA, Davis JP, Schloss PD, Bailey VL, Krumholz LR. Elshahed MS, et al. Appl Environ Microbiol. 2008 Sep;74(17):5422-8. doi: 10.1128/AEM.00410-08. Epub 2008 Jul 7. Appl Environ Microbiol. 2008. PMID: 18606799 Free PMC article.
References
- Abulencia, C. B., D. L. Wyborski, J. A. Garcia, M. Podar, W. Chen, S. H. Chang, H. W. Chang, D. Watson, E. L. Brodie, T. C. Hazen, and M. Keller. 2006. Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl. Environ. Microbiol. 72:3291-3301. - PMC - PubMed
- Baron, C. 2005. From bioremediation to biowarfare: on the impact and mechanism of type IV secretion systems. FEMS Microbiol. Lett. 253:163-170. - PubMed
- Beja, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. Delong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. - PubMed
- Berry, A. E., C. Chiocchini, T. Selby, M. Sosio, and E. M. Wellington. 2003. Isolation of high molecular weight DNA from soil for cloning into BAC vectors. FEMS Microbiol. Lett. 223:15-20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases