TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture - PubMed (original) (raw)
TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture
Lucio Conti et al. Plant Cell. 2007 Mar.
Abstract
Shoot meristems harbor stem cells that provide key growing points in plants, maintaining themselves and generating all above-ground tissues. Cell-to-cell signaling networks maintain this population, but how are meristem and organ identities controlled? TERMINAL FLOWER1 (TFL1) controls shoot meristem identity throughout the plant life cycle, affecting the number and identity of all above-ground organs generated; tfl1 mutant shoot meristems make fewer leaves, shoots, and flowers and change identity to flowers. We find that TFL1 mRNA is broadly distributed in young axillary shoot meristems but later becomes limited to central regions, yet affects cell fates at a distance. How is this achieved? We reveal that the TFL1 protein is a mobile signal that becomes evenly distributed across the meristem. TFL1 does not enter cells arising from the flanks of the meristem, thus allowing primordia to establish their identity. Surprisingly, TFL1 movement does not appear to occur in mature shoots of leafy (lfy) mutants, which eventually stop proliferating and convert to carpel/floral-like structures. We propose that signals from LFY in floral meristems may feed back to promote TFL1 protein movement in the shoot meristem. This novel feedback signaling mechanism would ensure that shoot meristem identity is maintained and the appropriate inflorescence architecture develops.
Figures
Figure 1.
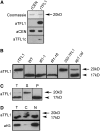
TFL1 Is a Cytoplasmic 20-kD Protein of Inflorescences. (A) Recombinant (r) CEN and TFL1 proteins (100 ng) were size-fractioned by SDS-PAGE and visualized with Coomassie staining to reveal the predicted size of 20 kD. Equivalent gels were blotted and probed with anti-TFL1 sera (aTFL1), anti-CEN sera (aCEN), or commercial anti-TFL1 sera (aTFL1c). (B) Anti-TFL1 sera were used to probe blots of rTFL1 compared with plant extracts. Total protein extracts were analyzed from inflorescences (1-cm-long shoots) of 16-d-old wild-type, tfl1-1, and tfl1-18 plants. Extracts of 12-d-old vegetative 35S-TFL1 seedlings and 26-d-old ap1 cal shoot meristem tissues were also analyzed. The arrow indicates the 20-kD TFL1 protein. The arrowhead indicates a nonspecific protein of 17 kD detected by the sera. (C) Total (T) protein extracts derived from ap1 cal meristem tissues were fractionated into a high-speed supernatant (S) fraction and membrane pellet (P). Proteins were blotted and probed with anti-TFL1 sera. (D) Total (T) protein extracts derived from ap1 cal meristem tissues were fractionated into a cytosolic (C) and nuclear fraction (N). Equal amounts of proteins were analyzed as in (C) and probed with anti-TFL1 sera (aTFL1). An equivalent blot was probed with anti-histone3 (aH3) sera.
Figure 2.
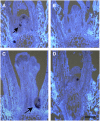
TFL1 mRNA Patterns in Shoot Inflorescence Meristems. (A) TFL1 mRNA pattern (purple stain) in sections of wild-type plants harvested after 12 LD. Strong TFL1 mRNA was detected in the main shoot and young axillary meristems (arrow). The tissue was counterstained to show cells (white). (B) No TFL1 mRNA was seen in any tfl1-18 meristems (10 LD is shown). (C) and (D) TFL1 expression was seen in 12-d-old tfl1-1 and tfl1-13 mutant plants in axillary meristems (arrow). Bar = 100 μm.
Figure 3.
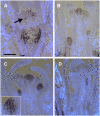
TFL1 Protein Moves beyond Its mRNA Domain. (A) and (B) TFL1 protein in wild-type (Col) plants harvested after 12 or 16 LD, respectively. TFL1 protein (purple stain) was detected at 12 LD in young axillary shoot meristems (arrow). At 16 LD, TFL1 protein was detected also in the main shoot meristem, its upper axillary meristems, and axillary shoot meristems of rosette leaves. The tissue was counterstained to show cells (white). Bar = 100 μm. (C) TFL1 protein was also detected in wild-type L_er_ plants harvested after 14 LD. Inset shows higher magnification (×3) of a meristem, highlighting TFL1 in epidermis/L1. (D) TFL1 protein was not detected in tfl1-18 mutant plants harvested after any time point (12 LD is shown).
Figure 4.
TFL1 mRNA in SD Vegetative Axillary Meristems. (A) and (B) TFL1 mRNA expression patterns in sections of wild-type plants grown for 30 SD (A) and then transferred to inductive LD and harvested after 4 LD (B). Bar = 100 μm. (C) and (D) tfl1-18 mRNA expression patterns in tfl1-18 mutant plants after 30 SD ([C]; inset shows that axillary meristems can have weak tfl1-18 mRNA at low frequency) and then after 2 LD induction (D). A section from 2 LD probed for AP1 expression (see Supplemental Figure 4 online) showed that many shoot meristems were present (arrows in [D]). (E) and (F) tfl1-1 mRNA expression pattern in tfl1-1 mutant plants after 30 SD (E) or after 4 LD induction (F).
Figure 5.
TFL1 Protein Moves in Vegetative Meristems. (A) to (D) TFL1 protein in wild-type plants grown for 30 SD (A) or after 3, 5, or 6 LD induction, respectively ([B] to [D]). Axillary meristems with strong TFL1 protein are marked in (C) with asterisks. (E) and (F) TFL1 protein pattern (E) was directly compared with its TFL1 mRNA pattern (F) in sections of the same material harvested after 30 SD + 5 LD. Again, TFL1 mRNA was restricted, while TFL1 protein was throughout the shoot meristems. (G) and (H) No mutant protein was detected in tfl1-18 mutant plants after 30 SD (G), after 1 LD induction (H), or any other times. Dark spots are dirt on tissue/slide. (I) Similarly, no mutant protein was detected in tfl1-1 mutant plants at any time point (30 SD + 2 LD is shown) despite abundant mRNA. Bar = 100 μm.
Figure 6.
TFL1 Protein Patterns in lfy and ap1 cal Mutants. (A) to (D) Analysis of TFL1 protein in lfy mutants ([A] to [C]) and a wild-type segregant (D). The lfy-6 allele was analyzed after 23 LD ([A], main shoot; inset shows secondary shoot with young axillary shoot meristem) or after 27 LD (B). The lfy-14 allele was analyzed after 27 LD (C) and compared with the wild type (D). (E) to (H) TFL1 protein was detected in ap1 cal mutant shoot meristems after 12, 14, 16, and 20 LD, respectively. Bar = 100 μm.
Figure 7.
Model for TFL1 Expression, Movement, Regulation, and Action. TFL1 mRNA is first found throughout all cells of young axillary meristems in the axils of leaves. As the meristem develops, TFL1 mRNA is excluded from the tip, but TFL1 protein moves into this region to coordinate cell identity. In the mature shoot meristem, TFL1 mRNA becomes limited to central cells, while TFL1 protein continues to move to the outer cells (black arrows). In the inflorescence, LFY is expressed in peripheral cells (anlagen) and floral meristems, and signals (gray arrows) from LFY promote TFL1 protein movement in the shoot meristem and thus restrict LFY and AP1 expression.
Similar articles
- Changing the spatial pattern of TFL1 expression reveals its key role in the shoot meristem in controlling Arabidopsis flowering architecture.
Baumann K, Venail J, Berbel A, Domenech MJ, Money T, Conti L, Hanzawa Y, Madueno F, Bradley D. Baumann K, et al. J Exp Bot. 2015 Aug;66(15):4769-80. doi: 10.1093/jxb/erv247. Epub 2015 May 27. J Exp Bot. 2015. PMID: 26019254 Free PMC article. - Separate elements of the TERMINAL FLOWER 1 cis-regulatory region integrate pathways to control flowering time and shoot meristem identity.
Serrano-Mislata A, Fernández-Nohales P, Doménech MJ, Hanzawa Y, Bradley D, Madueño F. Serrano-Mislata A, et al. Development. 2016 Sep 15;143(18):3315-27. doi: 10.1242/dev.135269. Epub 2016 Jul 6. Development. 2016. PMID: 27385013 - Genetic interactions reveal the antagonistic roles of FT/TSF and TFL1 in the determination of inflorescence meristem identity in Arabidopsis.
Lee C, Kim SJ, Jin S, Susila H, Youn G, Nasim Z, Alavilli H, Chung KS, Yoo SJ, Ahn JH. Lee C, et al. Plant J. 2019 Aug;99(3):452-464. doi: 10.1111/tpj.14335. Epub 2019 May 17. Plant J. 2019. PMID: 30943325 - Turning Meristems into Fortresses.
Périlleux C, Bouché F, Randoux M, Orman-Ligeza B. Périlleux C, et al. Trends Plant Sci. 2019 May;24(5):431-442. doi: 10.1016/j.tplants.2019.02.004. Epub 2019 Mar 7. Trends Plant Sci. 2019. PMID: 30853243 Review. - Coming into bloom: the specification of floral meristems.
Liu C, Thong Z, Yu H. Liu C, et al. Development. 2009 Oct;136(20):3379-91. doi: 10.1242/dev.033076. Development. 2009. PMID: 19783733 Review.
Cited by
- Mobile TERMINAL FLOWER1 determines seed size in Arabidopsis.
Zhang B, Li C, Li Y, Yu H. Zhang B, et al. Nat Plants. 2020 Sep;6(9):1146-1157. doi: 10.1038/s41477-020-0749-5. Epub 2020 Aug 24. Nat Plants. 2020. PMID: 32839516 - Characterization and Functional Analysis of PEBP Family Genes in Upland Cotton (Gossypium hirsutum L.).
Zhang X, Wang C, Pang C, Wei H, Wang H, Song M, Fan S, Yu S. Zhang X, et al. PLoS One. 2016 Aug 23;11(8):e0161080. doi: 10.1371/journal.pone.0161080. eCollection 2016. PLoS One. 2016. PMID: 27552108 Free PMC article. - CsTFL1 inhibits determinate growth and terminal flower formation through interaction with CsNOT2a in cucumber.
Wen C, Zhao W, Liu W, Yang L, Wang Y, Liu X, Xu Y, Ren H, Guo Y, Li C, Li J, Weng Y, Zhang X. Wen C, et al. Development. 2019 Jul 29;146(14):dev180166. doi: 10.1242/dev.180166. Development. 2019. PMID: 31320327 Free PMC article. - Comparative genomics of flowering behavior in Cannabis sativa.
Steel L, Welling M, Ristevski N, Johnson K, Gendall A. Steel L, et al. Front Plant Sci. 2023 Jul 27;14:1227898. doi: 10.3389/fpls.2023.1227898. eCollection 2023. Front Plant Sci. 2023. PMID: 37575928 Free PMC article. - Plant Inflorescence Architecture: The Formation, Activity, and Fate of Axillary Meristems.
Zhu Y, Wagner D. Zhu Y, et al. Cold Spring Harb Perspect Biol. 2020 Jan 2;12(1):a034652. doi: 10.1101/cshperspect.a034652. Cold Spring Harb Perspect Biol. 2020. PMID: 31308142 Free PMC article. Review.
References
- Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056. - PubMed
- Alvarez, J., Guli, C.L., Yu, X.-H., and Smyth, D.R. (1992). terminal flower: A gene affecting inflorescence development in Arabidopsis thaliana. Plant J. 2 103–116.
- An, H., Roussot, C., Suarez-Lopez, P., Corbesier, L., Vincent, C., Pineiro, M., Hepworth, S., Mouradov, A., Justin, S., Turnbull, C., and Coupland, G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131 3615–3626. - PubMed
- Banfield, M.J., and Brady, R.L. (2000). The structure of Antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J. Mol. Biol. 297 1159–1170. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases