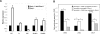Critical roles for Dicer in the female germline - PubMed (original) (raw)
Critical roles for Dicer in the female germline
Elizabeth P Murchison et al. Genes Dev. 2007.
Abstract
Dicer is an essential component of RNA interference (RNAi) pathways, which have broad functions in gene regulation and genome organization. Probing the consequences of tissue-restricted Dicer loss in mice indicates a critical role for Dicer during meiosis in the female germline. Mouse oocytes lacking Dicer arrest in meiosis I with multiple disorganized spindles and severe chromosome congression defects. Oogenesis and early development are times of significant post-transcriptional regulation, with controlled mRNA storage, translation, and degradation. Our results suggest that Dicer is essential for turnover of a substantial subset of maternal transcripts that are normally lost during oocyte maturation. Furthermore, we find evidence that transposon-derived sequence elements may contribute to the metabolism of maternal transcripts through a Dicer-dependent pathway. Our studies identify Dicer as central to a regulatory network that controls oocyte gene expression programs and that promotes genomic integrity in a cell type notoriously susceptible to aneuploidy.
Figures
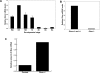
Figure 1.
Dicer is expressed in growing oocytes, and is depleted in oocytes from Dicerflox/flox; Zp3-Cre mice. (A) Temporal pattern of Dicer mRNA expression. Quantitative RT–PCR was performed in mouse oocytes and preimplantation embryos. Metaphase II eggs; one-cell, two-cell, eight-cell embryos; and blastocysts were harvested 13 h, 20 h, 44 h, 68 h, and 96 h post-hCG, respectively. The values are normalized to the amount of Dicer mRNA in GV oocytes. The experiment was performed three times, and the data are presented as the mean ± SEM. (INC) Meiotically incompetent oocyte; (GV) fully grown GV-intact oocyte; (MII) metaphase II-arrested egg; (1C) one-cell stage embryo; (2C) two-cell stage embryo; (8C) eight-cell stage embryo; (BL) blastocyst. (B) Dicer mRNA levels in wild-type and mutant oocytes. Quantitative real-time PCR was performed. The experiment was performed five times, and the data are presented as the mean ± SEM. (C) Dicer is functionally depleted in Dicerflox/flox; Zp3-Cre oocytes. Twenty-four hours after Mos dsRNA injection into Dicerflox/flox; Zp3-Cre or control oocytes, Mos mRNA levels were measured by quantitative RT–PCR.
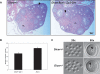
Figure 2.
Dicer is dispensable for oocyte growth and hormone response. (A) Ovarian histology in Dicerflox/flox; Zp3-Cre or wild-type (Dicer+/−) female mice. The ovaries were fixed overnight in Bouin’s fixative, embedded in paraffin, and sliced into 10-μm sections and stained with hematoxylin and eosin. The arrowheads indicate secondary follicles. (CL) Corpus luteum. (B) Number of oocytes recovered from Dicerflox/flox; Zp3-Cre (Dicer−/−) or wild-type (Dicer+/+ and Dicer+/−) female mice. Fully grown GV-intact oocytes were collected from the ovaries of PMSG-primed mice as described in Materials and Methods. The data are presented as the mean ± SEM; 19 wild-type and 12 mutant mice were analyzed. The difference between the two groups is not statistically significant (Student’s two-tailed _t_-test, p = 0.05). (C) Morphology of Dicer+/− and Dicer−/− oocytes. Oocytes were collected as described and bright-field microscopy was performed. Arrows indicate the prominent nucleoli characteristic of GV oocytes.
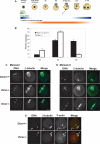
Figure 3.
Dicer is required for completion of meiotic maturation. (A) Schematic of oocyte maturation and early embryo development. The transition from fully grown GV oocyte to the late one-cell embryo occurs in the absence of significant transcription and relies on translation of maternal mRNA transcripts. The first part of this transition, when an oocyte completes meiosis I and arrests at metaphase of meiosis II is known as meiotic maturation. Zp3-Cre is expressed only in growing oocytes. (B) The maturation stage of Dicer−/− and control oocytes was scored by the presence of a GV (GV oocyte), a polar body (MII oocyte), or neither a GV nor a polar body (MI oocyte). Six-hundred-ninety-four Dicer+/+ and Dicer+/− and 582 Dicer−/− oocytes were analyzed in total. Error bars denote standard error of the mean. (C,D) Spindle morphology in Dicer−/− oocytes matured in vitro to metaphase I (C) or metaphase II (D). Oocytes were fixed in 3.7% paraformaldehyde and stained with a β-tubulin antibody; DNA was counterstained with DAPI or Sytox green. Representative images are shown. Bar, 25 μm. (E) Cortical actin and polar body morphology in mutant oocytes. Oocytes were fixed in 3.7% paraformaldehyde and stained with a β-tubulin antibody; F-actin was labeled with phalloidin-Alexa fluor 635 and DNA was counterstained with DAPI. The arrowheads indicate the position of the first polar body. β-Tubulin is often present inside the polar body in both wild-type and mutant oocytes.
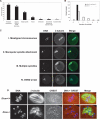
Figure 4.
Loss of Dicer causes a range of spindle and chromosome congression defects. (A) Frequency distribution of Dicer−/− oocyte phenotypes. The absolute number of oocytes observed with a given phenotype is indicated above the bar. One oocyte may fall into more than one phenotype class. (B) Quantification of number of spindles per oocyte in Dicer−/− and wild-type oocytes. (C) β-Tubulin immunofluorescence staining of Dicer−/− oocytes exhibiting several common phenotypes. DNA was counterstained with DAPI or Sytox green. (D) Centromere staining in Dicer−/− oocytes. Oocytes were fixed in 3.7% paraformaldehyde and stained with a CREST antiserum together with a β-tubulin antibody; DNA was counterstained with DAPI.
Figure 5.
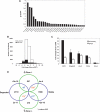
miRNAs and siRNAs may accelerate degradation of maternal transcripts. (A) Relative expression levels of the 30 most abundantly detected miRNAs in GV oocytes (out of a set of 103 tested miRNAs). miRNAs were quantified as described in Materials and Methods. Z-score expresses the divergence of the experimental result from the most probable result (mean) as a number of standard deviations. The larger the value of Z, the less probable the experimental result is due to chance. (B) Distribution of probe changes in the microarray comparing Dicer−/− to wild-type oocytes. FDR ≤5% (shaded region) denotes the probes whose changes hold greatest confidence. (C) Confirmation of selected microarray changes in Dicer−/− oocytes by quantitative RT–PCR. Fold change between Dicer−/− and wild-type oocytes is indicated on the _Y_-axis. Error bars denote standard error of the mean. (D) Superposition of probes that change in Dicer−/− with those that are degraded during meiotic maturation (Su et al. 2006). Only probe sets called present in both experiments (19,301 probe sets) were used in this analysis. Green circles show the number of probe sets significantly changed at least twofold in Dicer−/− oocytes, and violet circles show the probe sets degraded or kept stable from GV to MII (Su et al. 2006). The overlapping probe sets are shown in the overlapping region, together with the _p_-value by χ2 test. Statistical significance (_p_-value < 0.05) is indicated by an asterisk (*). Blue indicates a negative correlation between overlapping probe sets and red means two sets are positively correlated.
Figure 6.
Transposons and mRNAs harboring transposon-derived sequences are misregulated in Dicer−/− oocytes. (A) Quantitation of retrotransposon levels in Dicer−/− oocytes by RT–PCR. Error bars indicate standard error of the mean. (B) Quantitation of repeat element representation in 3′UTRs of transcripts that are either up-regulated, down-regulated, or remain stable in the absence of Dicer. Values have been normalized to the percent 3′UTR repeat content of the unchanged set. Asterisk (*)denotes statistical significance (p < 1.0e−10) of repeat proportion difference (percent of 3′UTR contributed by repetitive elements) between two sets of transcripts calculated by prop.test in R package (
).
Similar articles
- Functional genomic analysis identifies miRNA repertoire regulating C. elegans oocyte development.
Minogue AL, Tackett MR, Atabakhsh E, Tejada G, Arur S. Minogue AL, et al. Nat Commun. 2018 Dec 14;9(1):5318. doi: 10.1038/s41467-018-07791-w. Nat Commun. 2018. PMID: 30552320 Free PMC article. - Maternal microRNAs are essential for mouse zygotic development.
Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Tang F, et al. Genes Dev. 2007 Mar 15;21(6):644-8. doi: 10.1101/gad.418707. Genes Dev. 2007. PMID: 17369397 Free PMC article. - Dicer-1 is a key enzyme in the regulation of oogenesis in panoistic ovaries.
Tanaka ED, Piulachs MD. Tanaka ED, et al. Biol Cell. 2012 Aug;104(8):452-61. doi: 10.1111/boc.201100044. Epub 2012 May 18. Biol Cell. 2012. PMID: 22462497 - [Advance on Dicer gene and its role in female reproduction].
Li P, Zhu WJ. Li P, et al. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2011 Jun;28(3):275-8. doi: 10.3760/cma.j.issn.1003-9406.2011.03.008. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2011. PMID: 21644222 Review. Chinese. - Role of Dicer in female fertility.
Luense LJ, Carletti MZ, Christenson LK. Luense LJ, et al. Trends Endocrinol Metab. 2009 Aug;20(6):265-72. doi: 10.1016/j.tem.2009.05.001. Epub 2009 Jul 29. Trends Endocrinol Metab. 2009. PMID: 19646895 Free PMC article. Review.
Cited by
- Understanding transgenerational epigenetic inheritance via the gametes in mammals.
Daxinger L, Whitelaw E. Daxinger L, et al. Nat Rev Genet. 2012 Jan 31;13(3):153-62. doi: 10.1038/nrg3188. Nat Rev Genet. 2012. PMID: 22290458 Review. - Differential expression of dicer, miRNAs, and inflammatory markers in diabetic Ins2+/- Akita hearts.
Chavali V, Tyagi SC, Mishra PK. Chavali V, et al. Cell Biochem Biophys. 2014 Jan;68(1):25-35. doi: 10.1007/s12013-013-9679-4. Cell Biochem Biophys. 2014. PMID: 23797610 Free PMC article. - miRNAs as modulators of angiogenesis.
Landskroner-Eiger S, Moneke I, Sessa WC. Landskroner-Eiger S, et al. Cold Spring Harb Perspect Med. 2013 Feb 1;3(2):a006643. doi: 10.1101/cshperspect.a006643. Cold Spring Harb Perspect Med. 2013. PMID: 23169571 Free PMC article. Review. - Essential role for Argonaute2 protein in mouse oogenesis.
Kaneda M, Tang F, O'Carroll D, Lao K, Surani MA. Kaneda M, et al. Epigenetics Chromatin. 2009 Aug 10;2(1):9. doi: 10.1186/1756-8935-2-9. Epigenetics Chromatin. 2009. PMID: 19664249 Free PMC article. - The regulatory role of Dicer in folliculogenesis in mice.
Lei L, Jin S, Gonzalez G, Behringer RR, Woodruff TK. Lei L, et al. Mol Cell Endocrinol. 2010 Feb 5;315(1-2):63-73. doi: 10.1016/j.mce.2009.09.021. Epub 2009 Sep 30. Mol Cell Endocrinol. 2010. PMID: 19799966 Free PMC article.
References
- Amanai M., Brahmajosyula M., Perry A.C., Brahmajosyula M., Perry A.C., Perry A.C. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol. Reprod. 2006;75:877–884. - PubMed
- Andl T., Murchison E.P., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Murchison E.P., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Zhang Y., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Seykora J.T., Hannon G.J., Millar S.E., Hannon G.J., Millar S.E., Millar S.E. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 2006;16:1041–1049. - PMC - PubMed
- Anger M., Stein P., Schultz R.M., Stein P., Schultz R.M., Schultz R.M. CDC6 requirement for spindle formation during maturation of mouse oocytes. Biol. Reprod. 2005;72:188–194. - PubMed
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovina N., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovina N., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovina N., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Lagos-Quintana M., Landgraf P., Iovina N., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Landgraf P., Iovina N., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Iovina N., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Kuramouchi-Miyaqawa S., Nakano T., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HG001696/HG/NHGRI NIH HHS/United States
- R01 HD022681/HD/NICHD NIH HHS/United States
- HD22681/HD/NICHD NIH HHS/United States
- R01 HG001696/HG/NHGRI NIH HHS/United States
- R37 HD022681/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases