Advances in QTL mapping in pigs - PubMed (original) (raw)
Review
Advances in QTL mapping in pigs
Max F Rothschild et al. Int J Biol Sci. 2007.
Abstract
Over the past 15 years advances in the porcine genetic linkage map and discovery of useful candidate genes have led to valuable gene and trait information being discovered. Early use of exotic breed crosses and now commercial breed crosses for quantitative trait loci (QTL) scans and candidate gene analyses have led to 110 publications which have identified 1,675 QTL. Additionally, these studies continue to identify genes associated with economically important traits such as growth rate, leanness, feed intake, meat quality, litter size, and disease resistance. A well developed QTL database called PigQTLdb is now as a valuable tool for summarizing and pinpointing in silico regions of interest to researchers. The commercial pig industry is actively incorporating these markers in marker-assisted selection along with traditional performance information to improve traits of economic performance. The long awaited sequencing efforts are also now beginning to provide sequence available for both comparative genomics and large scale single nucleotide polymorphism (SNP) association studies. While these advances are all positive, development of useful new trait families and measurement of new or underlying traits still limits future discoveries. A review of these developments is presented.
Conflict of interest statement
Conflict of interest: The authors have declared that no conflict of interest exists.
Figures
Figure 1
Front page of the Pig QTLdb (
http://www.animalgenome.org/QTLdb/pig.html
), showing database summaries and ways the database may be accessed.
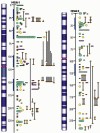
Figure 2
Example of mammalian concordant lipogenesis QTL maps for human chromosomes 1 and 2 (HSA1 and HSA2) as references according to number of obesity/fat QTL reported in pig (purple), human (green), mouse (yellow), and cow (orange). The blue bars indicate the putative concordant fat QTL regions.
Similar articles
- Cracking the genomic piggy bank: identifying secrets of the pig genome.
Mote BE, Rothschild MF. Mote BE, et al. Genome Dyn. 2006;2:86-96. doi: 10.1159/000095097. Genome Dyn. 2006. PMID: 18753772 Review. - Porcine genomics delivers new tools and results: this little piggy did more than just go to market.
Rothschild MF. Rothschild MF. Genet Res. 2004 Feb;83(1):1-6. doi: 10.1017/s0016672303006621. Genet Res. 2004. PMID: 15125061 Review. - A QTL resource and comparison tool for pigs: PigQTLDB.
Hu ZL, Dracheva S, Jang W, Maglott D, Bastiaansen J, Rothschild MF, Reecy JM. Hu ZL, et al. Mamm Genome. 2005 Oct;16(10):792-800. doi: 10.1007/s00335-005-0060-9. Epub 2005 Oct 29. Mamm Genome. 2005. PMID: 16261421 - Investigation of QTL regions on Chromosome 17 for genes associated with meat color in the pig.
Fan B, Glenn KL, Geiger B, Mileham A, Rothschild MF. Fan B, et al. J Anim Breed Genet. 2008 Aug;125(4):240-7. doi: 10.1111/j.1439-0388.2008.00749.x. J Anim Breed Genet. 2008. PMID: 18717966 - From a sow's ear to a silk purse: real progress in porcine genomics.
Rothschild MF. Rothschild MF. Cytogenet Genome Res. 2003;102(1-4):95-9. doi: 10.1159/000075732. Cytogenet Genome Res. 2003. PMID: 14970686 Review.
Cited by
- A Whole Genome Association Study on Meat Quality Traits Using High Density SNP Chips in a Cross between Korean Native Pig and Landrace.
Lee KT, Lee YM, Alam M, Choi BH, Park MR, Kim KS, Kim TH, Kim JJ. Lee KT, et al. Asian-Australas J Anim Sci. 2012 Nov;25(11):1529-39. doi: 10.5713/ajas.2012.12474. Asian-Australas J Anim Sci. 2012. PMID: 25049513 Free PMC article. - Power and precision of QTL mapping in simulated multiple porcine F2 crosses using whole-genome sequence information.
Schmid M, Wellmann R, Bennewitz J. Schmid M, et al. BMC Genet. 2018 Apr 3;19(1):22. doi: 10.1186/s12863-018-0604-0. BMC Genet. 2018. PMID: 29614956 Free PMC article. - AGBE: a dual deaminase-mediated base editor by fusing CGBE with ABE for creating a saturated mutant population with multiple editing patterns.
Liang Y, Xie J, Zhang Q, Wang X, Gou S, Lin L, Chen T, Ge W, Zhuang Z, Lian M, Chen F, Li N, Ouyang Z, Lai C, Liu X, Li L, Ye Y, Wu H, Wang K, Lai L. Liang Y, et al. Nucleic Acids Res. 2022 May 20;50(9):5384-5399. doi: 10.1093/nar/gkac353. Nucleic Acids Res. 2022. PMID: 35544322 Free PMC article. - Obesity gene atlas in mammals.
Kunej T, Jevsinek Skok D, Zorc M, Ogrinc A, Michal JJ, Kovac M, Jiang Z. Kunej T, et al. J Genomics. 2013 Dec 1;1:45-55. doi: 10.7150/jgen.3996. eCollection 2013. J Genomics. 2013. PMID: 25031655 Free PMC article. - QTL fine mapping with Bayes C(π): a simulation study.
van den Berg I, Fritz S, Boichard D. van den Berg I, et al. Genet Sel Evol. 2013 Jun 19;45(1):19. doi: 10.1186/1297-9686-45-19. Genet Sel Evol. 2013. PMID: 23782975 Free PMC article.
References
- Rothschild MF, Ruvinsky A. Genetics of the Pig. Oxon, UK: CABI Press; 1998.
- Mason IL. A World Dictionary of Livestock Breeds, Types, and Varieties. 2nd Edition. Farnham Royal, UK: CAB International; 1969.
- Archibald AL, Brown JF, Couperwhite S et al. The PiGMaP consortium linkage map of the pig (Sus scrofa) Mammalian Genome. 1995;6:157–175. - PubMed
- Rohrer GA, Alexander LJ, Hu ZL et al. A comprehensive map of the porcine genome. Genome Research. 1996;6:371–391. - PubMed
- Mote B, Rothschild MF. Cracking the Genomic Piggy Bank: Identifying Secrets of the Pig Genome. Genome Dynamics. 2006;2:86–96. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous