Destruction of long-range interactions by a single mutation in lysozyme - PubMed (original) (raw)
Comparative Study
. 2007 Apr 3;104(14):5824-9.
doi: 10.1073/pnas.0701249104. Epub 2007 Mar 26.
Affiliations
- PMID: 17389393
- PMCID: PMC1851576
- DOI: 10.1073/pnas.0701249104
Comparative Study
Destruction of long-range interactions by a single mutation in lysozyme
Ruhong Zhou et al. Proc Natl Acad Sci U S A. 2007.
Erratum in
- Proc Natl Acad Sci U S A. 2007 Jul 3;104(27):11507
Abstract
We propose a mechanism, based on a > or =10-micros molecular dynamics simulation, for the surprising misfolding of hen egg-white lysozyme caused by a single mutation (W62G). Our simulations of the wild-type and mutant lysozymes in 8 M urea solution at biological temperature (with both pH 2 and 7) reveal that the mutant structure is much less stable than that of the wild type, with the mutant showing larger fluctuations and less native-like contacts. Analysis of local contacts reveals that the Trp-62 residue is the key to a cooperative long-range interaction within the wild type, where it acts like a bridge between two neighboring basic residues. Thus, a native-like cluster or nucleation site can form near these residues in the wild type but not in the mutant. The time evolution of the secondary structure also exhibits a quicker loss of the beta-sheets in the mutant than in the wild type, whereas some of the alpha-helices persist during the entire simulation in both the wild type and the mutant in 8 M urea (even though the tertiary structures are basically all gone). These findings, while supporting the general conclusions of a recent experimental study by Dobson and coworkers [Klein-Seetharam J, Oikama M, Grimshaw SB, Wirmer J, Duchardt E, Ueda T, Imoto T, Smith LJ, Dobson CM, Schwalbe H (2002) Science 295:1719-1722], provide a detailed but different molecular picture of the misfolding mechanism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
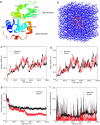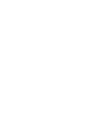
Fig. 1.
System and simulation results. (a) Ribbon view of the native lysozyme protein, with residue Trp-62 represented in sticks and both α- and β-domains marked. (b) Solvated lysozyme in 8 M urea solution, with protein lysozyme represented as red ribbons, urea as blue sticks, and water as white sticks. (c) Comparison of the backbone rmsd for the wild-type and mutant lysozymes. (d) Comparison of the radius gyration. (e) Comparison of fraction of native contacts. (f) Comparison of local contact of the mutation site (residue 62).
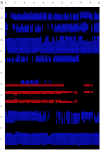
Fig. 2.
Time evolution of the secondary structure at 0- to 100-ns MD simulation for the wild type (a) and W62G mutant (b). The secondary structure is assigned by the program STRIDE (19), with the α-helix colored blue, the 310-helix light blue, the β-strand red, and the coils and turns black. The secondary structure of the starting crystal structure is displayed at t = 0 ns [helix A (5–14), helix B (25–36), helix C (90–100), helix D (110–115), strand 1 (43–46), strand 2 (51–54), and a 310-helix (81–85)].
Fig. 2.
Time evolution of the secondary structure at 0- to 100-ns MD simulation for the wild type (a) and W62G mutant (b). The secondary structure is assigned by the program STRIDE (19), with the α-helix colored blue, the 310-helix light blue, the β-strand red, and the coils and turns black. The secondary structure of the starting crystal structure is displayed at t = 0 ns [helix A (5–14), helix B (25–36), helix C (90–100), helix D (110–115), strand 1 (43–46), strand 2 (51–54), and a 310-helix (81–85)].
Fig. 3.
Snapshots of the mutant lysozyme during one representative microsecond of the chemical denaturing trajectories. These snapshots clearly indicate the gradual loss of the native contacts, with most of the loss in the β-domain first. Interestingly, even at the end of the 1-μs simulation, some helical content still persists.
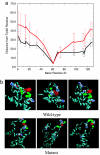
Fig. 4.
Trp-62 interactions with basic residues. (a) Comparison of the average distance and SD of basic residues from Trp-62 (Cα–Cα distance) for the wild type and mutant in the first 100-ns simulation. (b) Representative structures of the wild type and mutant during the 100- to 1,000-ns MD simulations. The Trp-62 is represented as red van der Waals space-fills, and three nearby basic residues (Arg-73, Lys-97, Arg-112) are represented as van der Waals space-fills. The green balls represent the residues making local contacts with Trp-62.
Similar articles
- Single-mutation-induced stability loss in protein lysozyme.
Ye L, Wu Z, Eleftheriou M, Zhou R. Ye L, et al. Biochem Soc Trans. 2007 Dec;35(Pt 6):1551-7. doi: 10.1042/BST0351551. Biochem Soc Trans. 2007. PMID: 18031265 - Thermal denaturing of mutant lysozyme with both the OPLSAA and the CHARMM force fields.
Eleftheriou M, Germain RS, Royyuru AK, Zhou R. Eleftheriou M, et al. J Am Chem Soc. 2006 Oct 18;128(41):13388-95. doi: 10.1021/ja060972s. J Am Chem Soc. 2006. PMID: 17031950 - Cooperative folding of the isolated alpha-helical domain of hen egg-white lysozyme.
Bai P, Peng Z. Bai P, et al. J Mol Biol. 2001 Nov 23;314(2):321-9. doi: 10.1006/jmbi.2001.5122. J Mol Biol. 2001. PMID: 11718563 - Folding of lysozyme.
Fischer B. Fischer B. EXS. 1996;75:143-61. doi: 10.1007/978-3-0348-9225-4_9. EXS. 1996. PMID: 8765299 Review.
Cited by
- Analysis of core region from egg white lysozyme forming amyloid fibrils.
Tokunaga Y, Sakakibara Y, Kamada Y, Watanabe K, Sugimoto Y. Tokunaga Y, et al. Int J Biol Sci. 2013;9(2):219-27. doi: 10.7150/ijbs.5380. Epub 2013 Feb 13. Int J Biol Sci. 2013. PMID: 23459392 Free PMC article. - Free-energy simulations reveal that both hydrophobic and polar interactions are important for influenza hemagglutinin antibody binding.
Xia Z, Huynh T, Kang SG, Zhou R. Xia Z, et al. Biophys J. 2012 Mar 21;102(6):1453-61. doi: 10.1016/j.bpj.2012.01.043. Epub 2012 Mar 20. Biophys J. 2012. PMID: 22455929 Free PMC article. - Comment on "urea-mediated protein denaturation: a consensus view".
Zhou R, Li J, Hua L, Yang Z, Berne BJ. Zhou R, et al. J Phys Chem B. 2011 Feb 10;115(5):1323-6; discussion 1327-8. doi: 10.1021/jp105160a. Epub 2011 Jan 19. J Phys Chem B. 2011. PMID: 21247088 Free PMC article. No abstract available. - Analyses of Mutation Displacements from Homology Models.
Carpentier M, Chomilier J. Carpentier M, et al. Methods Mol Biol. 2023;2627:195-210. doi: 10.1007/978-1-0716-2974-1_11. Methods Mol Biol. 2023. PMID: 36959449 - A novel 9-bp insertion detected in steroid 21-hydroxylase gene (CYP21A2): prediction of its structural and functional implications by computational methods.
Dubey S, Idicula-Thomas S, Anwaruddin M, Saravanan C, Varma RR, Maitra A. Dubey S, et al. J Biomed Sci. 2009 Jan 8;16(1):3. doi: 10.1186/1423-0127-16-3. J Biomed Sci. 2009. PMID: 19272182 Free PMC article.
References
- Kagan BL, Dobson CM. Science. 2005;307:42–43. - PubMed
- Klein-Seetharaman J, Oikawa M, Grimshaw SB, Wirmer J, Duchardt E, Ueda T, Imoto T, Smith LJ, Dobson CM, Schwalbe H. Science. 2002;295:1719–1722. - PubMed
- Dumoulin M, Last A, Desmyter A, Decanniere K, Canet D, Spencer A, Archer D, Muyldermans S, Wyns L, Matagne A, et al. Nature. 2003;424:783–788. - PubMed
- Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Nature. 2003;424:805–808. - PubMed
- Brooks CL, Onuchic JN, Wales DJ. Science. 2001;293:612–613. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources