Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions - PubMed (original) (raw)
Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions
Hongbo Xie et al. J Proteome Res. 2007 May.
Abstract
Identifying relationships between function, amino acid sequence, and protein structure represents a major challenge. In this study, we propose a bioinformatics approach that identifies functional keywords in the Swiss-Prot database that correlate with intrinsic disorder. A statistical evaluation is employed to rank the significance of these correlations. Protein sequence data redundancy and the relationship between protein length and protein structure were taken into consideration to ensure the quality of the statistical inferences. Over 200,000 proteins from the Swiss-Prot database were analyzed using this approach. The predictions of intrinsic disorder were carried out using PONDR VL3E predictor of long disordered regions that achieves an accuracy of above 86%. Overall, out of the 710 Swiss-Prot functional keywords that were each associated with at least 20 proteins, 238 were found to be strongly positively correlated with predicted long intrinsically disordered regions, whereas 302 were strongly negatively correlated with such regions. The remaining 170 keywords were ambiguous without strong positive or negative correlation with the disorder predictions. These functions cover a large variety of biological activities and imply that disordered regions are characterized by a wide functional repertoire. Our results agree well with literature findings, as we were able to find at least one illustrative example of functional disorder or order shown experimentally for the vast majority of keywords showing the strongest positive or negative correlation with intrinsic disorder. This work opens a series of three papers, which enriches the current view of protein structure-function relationships, especially with regards to functionalities of intrinsically disordered proteins, and provides researchers with a novel tool that could be used to improve the understanding of the relationships between protein structure and function. The first paper of the series describes our statistical approach, outlines the major findings, and provides illustrative examples of biological processes and functions positively and negatively correlated with intrinsic disorder.
Figures
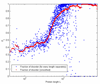
Figure 1
Fraction of putative disorder as a function of sequence length. The smoothed curve uses averaging window of size equal to 20% of the sequence length.
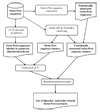
Figure 2
Schematic representation of the algorithm for extracting disorder- and order-related keywords.
Similar articles
- Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins.
Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Xie H, et al. J Proteome Res. 2007 May;6(5):1917-32. doi: 10.1021/pr060394e. Epub 2007 Mar 29. J Proteome Res. 2007. PMID: 17391016 Free PMC article. - Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions.
Vucetic S, Xie H, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Vucetic S, et al. J Proteome Res. 2007 May;6(5):1899-916. doi: 10.1021/pr060393m. Epub 2007 Mar 29. J Proteome Res. 2007. PMID: 17391015 Free PMC article. - Bioinformatics analysis of correlation between protein function and intrinsic disorder.
Vinterhalter G, Kovačević JJ, Uversky VN, Pavlović-Lažetić GM. Vinterhalter G, et al. Int J Biol Macromol. 2021 Jan 15;167:446-456. doi: 10.1016/j.ijbiomac.2020.11.211. Epub 2020 Dec 2. Int J Biol Macromol. 2021. PMID: 33278435 - Natively disordered proteins: functions and predictions.
Romero P, Obradovic Z, Dunker AK. Romero P, et al. Appl Bioinformatics. 2004;3(2-3):105-13. doi: 10.2165/00822942-200403020-00005. Appl Bioinformatics. 2004. PMID: 15693736 Review. - Intrinsic disorder and functional proteomics.
Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Radivojac P, et al. Biophys J. 2007 Mar 1;92(5):1439-56. doi: 10.1529/biophysj.106.094045. Epub 2006 Dec 8. Biophys J. 2007. PMID: 17158572 Free PMC article. Review.
Cited by
- Drugging the Undruggable: Targeting the N-Terminal Domain of Nuclear Hormone Receptors.
Sadar MD. Sadar MD. Adv Exp Med Biol. 2022;1390:311-326. doi: 10.1007/978-3-031-11836-4_18. Adv Exp Med Biol. 2022. PMID: 36107327 - A decade and a half of protein intrinsic disorder: biology still waits for physics.
Uversky VN. Uversky VN. Protein Sci. 2013 Jun;22(6):693-724. doi: 10.1002/pro.2261. Epub 2013 Apr 29. Protein Sci. 2013. PMID: 23553817 Free PMC article. Review. - A Phosphorylation-Induced Switch in the Nuclear Localization Sequence of the Intrinsically Disordered NUPR1 Hampers Binding to Importin.
Neira JL, Rizzuti B, Jiménez-Alesanco A, Palomino-Schätzlein M, Abián O, Velázquez-Campoy A, Iovanna JL. Neira JL, et al. Biomolecules. 2020 Sep 11;10(9):1313. doi: 10.3390/biom10091313. Biomolecules. 2020. PMID: 32933064 Free PMC article. - Rethinking gene regulatory networks in light of alternative splicing, intrinsically disordered protein domains, and post-translational modifications.
Niklas KJ, Bondos SE, Dunker AK, Newman SA. Niklas KJ, et al. Front Cell Dev Biol. 2015 Feb 26;3:8. doi: 10.3389/fcell.2015.00008. eCollection 2015. Front Cell Dev Biol. 2015. PMID: 25767796 Free PMC article. - An Emerging Role for Post-translational Modifications in Regulating RNP Condensates in the Germ Line.
Schisa JA, Elaswad MT. Schisa JA, et al. Front Mol Biosci. 2021 Apr 8;8:658020. doi: 10.3389/fmolb.2021.658020. eCollection 2021. Front Mol Biosci. 2021. PMID: 33898525 Free PMC article. Review.
References
- Fischer E. Einfluss der Configuration auf die Wirkung der Enzyme. Ber. Dt. Chem. Ges. 1894;27:2985–2993.
- Wu H. Studies on denaturation of proteins XIII A theory of denaturation. Chin. J. Physiol. 1931;1:219–234. - PubMed
- Sela M, White FH, Jr, Anfinsen CB. Reductive cleavage of disulfide bridges in ribonuclease. Science. 1957;125:691–692. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources