A subpopulation of human peripheral blood NK cells that lacks inhibitory receptors for self-MHC is developmentally immature - PubMed (original) (raw)
A subpopulation of human peripheral blood NK cells that lacks inhibitory receptors for self-MHC is developmentally immature
Sarah Cooley et al. Blood. 2007.
Abstract
How receptor acquisition correlates with the functional maturation of natural killer (NK) cells is poorly understood. We used quantitative real-time polymerase chain reaction (PCR) assays to compare NKG2 and killer immunoglobulin-like receptor (KIR) gene expression in NK cells from allogeneic transplant recipients and their donors. Marked differences were observed in the NK subsets of recipients who had 8-fold more CD56(bright) cells, diminished KIR expression (except 2DL4), and increased NKG2A. In normal blood not all CD56(dim) cells express KIR, and a novel subpopulation of cells committed to the NK-cell lineage was defined. These cells, which comprise 19.4% +/- 2.8% of the CD56(dim) NK population in healthy donors, express the activating NKG2D and NKG2E receptors but no KIR or NKG2A. Although the CD56(dim) NKG2A(-) KIR(-) NK cells lack "at least one" inhibitory receptor for autologous MHC class I, they are not fully responsive, but rather functionally immature cells with poor cytotoxicity and IFN-gamma production. Upon culture with IL-15 and a stromal cell line, CD56(dim) and CD56(bright) KIR(-) NK cells proliferate, express KIR, and develop cytotoxicity and cytokine-producing potential. These findings have implications for the function of NK cells reconstituting after transplantation and support a model for in vivo development in which CD56(bright) cells precede CD56(dim) cells.
Figures
Figure 1
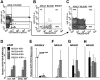
NK cells reconstituted 100 days after allogeneic transplantation are more frequently CD56bright and display a distinctive repertoire of NK-cell receptors compared with the donor's NK cells. (A) Comparison of NKR expression in blood mononuclear cells of recipients 100 days after transplantation and their donors, as assessed by Q-RT-PCR of 15 individual NKR genes. For each gene the mean recipient/donor ratio obtained from analysis of 32 transplant pairs is shown. Not all KIR were analyzed for all transplant pairs, because one or more KIR genes were absent from the donor KIR genotype. Variation in the number of NK cells in the mononuclear cell fractions was corrected by normalization of the NKR transcript numbers to the NCAM (CD56) transcript number. Error bars indicate standard error of the mean (SEM). (B-E) Flow cytometric analysis comparing the surface expression of KIR (B-C) and NKG2A (D-E) on CD56+CD3− gated NK cells from one representative transplant recipient (panels B,D) and the donor (panels C,E). The proportion of cells in each of the 4 quadrants is given as a percentage. The KIR analysis used a cocktail of the EB6, GL183, and DX9 mAbs.
Figure 2
NKG2A and KIR expression distinguishes populations of CD56dim NK cells. PBMCs were enriched for NK cells using a negative immunomagnetic bead depletion strategy. A representative example of a 4-color analysis shows gating of (A) CD56bright and CD56dim cells and analysis of the expression of NKG2A and KIR (using a cocktail of EB6, GL183, and DX9 mAbs) on (B) CD56bright and (C) CD56dim cells. Each of the 4 subpopulations shown in panel C was sorted and then analyzed by Q-RT-PCR for the expression of (D) KIR exhibiting variegated expression (vr) (n = 33 reactions), (E) KIR2DL4 (n = 6), (F) NKG2A (n = 7), (G) NKG2D (n = 7), and (H) NKG2E (n = 7). The numbers of NKG2 and KIR2DL4 transcripts in each subpopulation are compared with the positive control of IL-2 activated NK cells to give a relative expression. For comparison of vr_KIR_ expression in the 4 subpopulations, the data for each KIR were normalized to those of the NKG2A+KIR+ (designated by a †). These values were used to calculate a mean relative expression for the activating (KIR2DS1–3, 2DS5, 3DS1) and inhibitory (KIR2DL1–3, 2DL5, 3DL1, 3DL2). When a KIR was not expressed in the NKG2A+KIR+ population, indicating absence of the gene, it was excluded from the calculation of mean relative expression. Error bars indicate SEM.
Figure 3
NK-cell subsets differentially express surface antigens. PBMCs were stained with anti-CD56, anti-CD3, a cocktail of NKG2A and KIR antibodies, and FITC-conjugated antibodies against CD2, 16, 44, 62L, 94, and 161. NK-cell subsets were gated on the expression of CD56, and on the presence or absence of NKG2A and KIR. Shown are representative histograms (n = 5-11) for the surface antigens indicated. To facilitate comparison of antigen density, a vertical line (designated by a downward arrow above the CD56bright plots) is drawn through the approximate mean channel fluorescence of the subpopulation of CD56dim cells expressing NKG2A and/or KIR.
Figure 4
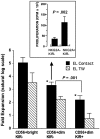
CD56dim NK cells proliferate less than CD56bright NK cells. Enriched NK cells were sorted into CD56bright KIR−, CD56dim KIR−, and CD56dim KIR+ NK cells and assayed for proliferation after 14 days of culture with the mouse stromal cell EL08–1D2 and exogenous IL-15 (n = 4). NK cells were either in direct contact with the stromal cells (black bars: EL Contact) or separated from them by a Transwell membrane (hatched bars: EL TW). These data are shown in the lower part of the figure: CD56bright KIR− NK cells proliferated more than either CD56dim subset (*P < .01) and the CD56dim KIR− population proliferated more than CD56dim KIR+ cells (P = .001). In the experiment shown in the boxed, upper part of the figure the CD56dim KIR− subset was further divided into CD56dim NKG2A−KIR− and NKG2A+KIR− subsets (n = 4) and tested in a 6-day thymidine incorporation assay where the NKG2A−KIR− subset showed less short-term responsiveness to IL-15 (10 ng/mL) (P = .002). Error bars indicate SEM.
Figure 5
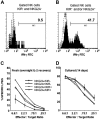
CD56 dim NK cells lacking NKG2A and KIR are functionally immature. PBMCs were incubated overnight with IL-12 and IL-18 and then stained with anti-CD56, anti-CD3, a cocktail of NKG2A and KIR antibodies, and IFN-γ for (A) CD56dim NKG2A−KIR− cells or (B) CD56dim NKG2A+ and/or KIR+ cells. Shown is a representative example of 12 experiments that all gave similar results. Sorted NKG2A−KIR−, NKG2A+KIR−, NKG2A−KIR+, and NKG2A+KIR+ populations were tested for cytolysis of K562 target cells in a 4-hour chromium release assay after (C) a 16-hour incubation with IL-2 (n = 6), to allow recovery of cytolytic machinery, or (D) after 14 days of maturation on EL08–1D2 and IL-15 (n = 4). After short-term culture with IL-2 the NKG2A−KIR− population exhibited a marked lack of cytolytic activity compared with the other populations (all P < .05). After maturation in culture for 14 days this difference was no longer apparent.
Figure 6
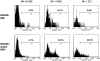
CD56dim NKG2A− KIR− cells are hyporesponsive to HLA class I–deficient target cells. Enriched NK cells were incubated alone, or after 4 hours of exposure to class I–deficient K562 or 221 cells. Cells were then stained for CD56, CD3, NKG2A, and KIR, and the degranulation antigen CD107a. Shown is a representative example gated on receptor-negative or -positive cells based on 6 experiments showing a similar pattern. The percents listed (M1 gate) represent positive events based on an isotype-matched control antibody.
Figure 7
CD56dim and CD56bright KIR− cells are precursors to receptor-expressing NK cells. CD56dim and CD56bright NK cells were sorted according to NKG2A and KIR expression, analyzed for NKR expression, cultured for 14 days on EL08–1D2 cells with IL-15, and then reanalyzed for NKR expression. (A) Shown is a representative analysis of the 4 subpopulations of CD56dim cells. The postsort analysis of each starting population (top row) is compared with similar analysis after culture to induce further maturation (bottom row). (B) Shown is an analysis of the CD56bright population in which essentially all cells were NKG2A+ (not shown). The postsort analysis (top) is compared with that after culture with EL08–1D2 cells and IL-15 (bottom).
Similar articles
- Killer Ig-like receptor (KIR) genotype predicts the capacity of human KIR-positive CD56dim NK cells to respond to pathogen-associated signals.
Korbel DS, Norman PJ, Newman KC, Horowitz A, Gendzekhadze K, Parham P, Riley EM. Korbel DS, et al. J Immunol. 2009 May 15;182(10):6426-34. doi: 10.4049/jimmunol.0804224. J Immunol. 2009. PMID: 19414796 Free PMC article. - NK cell killer Ig-like receptor repertoire acquisition and maturation are strongly modulated by HLA class I molecules.
Sleiman M, Brons NH, Kaoma T, Dogu F, Villa-Forte A, Lenoble P, Hentges F, Kotsch K, Gadola SD, Vilches C, Zimmer J. Sleiman M, et al. J Immunol. 2014 Mar 15;192(6):2602-10. doi: 10.4049/jimmunol.1302843. Epub 2014 Feb 19. J Immunol. 2014. PMID: 24554773 - Increased natural killer cell subsets with inhibitory cytokines and inhibitory surface receptors in patients with recurrent miscarriage and decreased or normal subsets in kidney transplant recipients late post-transplant.
Zhu L, Aly M, Wang H, Karakizlis H, Weimer R, Morath C, Kuon RJ, Toth B, Ekpoom N, Opelz G, Daniel V. Zhu L, et al. Clin Exp Immunol. 2018 Aug;193(2):241-254. doi: 10.1111/cei.13142. Epub 2018 May 31. Clin Exp Immunol. 2018. PMID: 29679490 Free PMC article. - Taking license with natural killer cell maturation and repertoire development.
Parham P. Parham P. Immunol Rev. 2006 Dec;214:155-60. doi: 10.1111/j.1600-065X.2006.00462.x. Immunol Rev. 2006. PMID: 17100883 Review. - Age-related changes in natural killer cell repertoires: impact on NK cell function and immune surveillance.
Manser AR, Uhrberg M. Manser AR, et al. Cancer Immunol Immunother. 2016 Apr;65(4):417-26. doi: 10.1007/s00262-015-1750-0. Epub 2015 Aug 19. Cancer Immunol Immunother. 2016. PMID: 26288343 Free PMC article. Review.
Cited by
- Comprehensive snapshots of natural killer cells functions, signaling, molecular mechanisms and clinical utilization.
Chen S, Zhu H, Jounaidi Y. Chen S, et al. Signal Transduct Target Ther. 2024 Nov 8;9(1):302. doi: 10.1038/s41392-024-02005-w. Signal Transduct Target Ther. 2024. PMID: 39511139 Free PMC article. Review. - Maintaining the Balance: Regulation of NK Cell Activity.
Imširović V, Wensveen FM, Polić B, Jelenčić V. Imširović V, et al. Cells. 2024 Aug 31;13(17):1464. doi: 10.3390/cells13171464. Cells. 2024. PMID: 39273034 Free PMC article. Review. - Genotype-specific expression of uncle fester suggests a role in allorecognition education in a basal chordate.
Taketa DA, Cengher L, Rodriguez D, Langenbacher AD, De Tomaso AW. Taketa DA, et al. bioRxiv [Preprint]. 2024 Feb 15:2024.02.13.580188. doi: 10.1101/2024.02.13.580188. bioRxiv. 2024. PMID: 38405917 Free PMC article. Updated. Preprint. - Anti-Inflammatory Effects of Nutritionally Relevant Concentrations of Oleuropein and Hydroxytyrosol on Peripheral Blood Mononuclear Cells: An Age-Related Analysis.
Pojero F, Gervasi F, Fiore SD, Aiello A, Bonacci S, Caldarella R, Attanzio A, Candore G, Caruso C, Ligotti ME, Procopio A, Restivo I, Tesoriere L, Allegra M, Accardi G. Pojero F, et al. Int J Mol Sci. 2023 Jul 3;24(13):11029. doi: 10.3390/ijms241311029. Int J Mol Sci. 2023. PMID: 37446206 Free PMC article. - Polymorphic KIR3DL3 expression modulates tissue-resident and innate-like T cells.
Palmer WH, Leaton LA, Campos Codo A, Crute B, Roest J, Zhu S, Petersen J, Tobin RP, Hume PS, Stone M, van Bokhoven A, Gerich ME, McCarter MD, Zhu Y, Janssen WJ, Vivian JP, Trowsdale J, Getahun A, Rossjohn J, Cambier J, Loh L, Norman PJ. Palmer WH, et al. Sci Immunol. 2023 Jun 30;8(84):eade5343. doi: 10.1126/sciimmunol.ade5343. Epub 2023 Jun 30. Sci Immunol. 2023. PMID: 37390222 Free PMC article.
References
- Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. - PubMed
- Raulet DH, Vance RE, McMahon CW. Regulation of the natural killer cell receptor repertoire. Annu Rev Immunol. 2001;19:291–330. - PubMed
- Valiante NM, Uhrberg M, Shilling HG, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. - PubMed
- Anfossi N, Andre P, Guia S, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA065493/CA/NCI NIH HHS/United States
- R01 HL055417/HL/NHLBI NIH HHS/United States
- K12 RR023247-01/RR/NCRR NIH HHS/United States
- R01-AI-22039/AI/NIAID NIH HHS/United States
- P01-CA-111412-01/CA/NCI NIH HHS/United States
- R01 AI022039/AI/NIAID NIH HHS/United States
- K12 RR023247/RR/NCRR NIH HHS/United States
- 240-97-0036/PHS HHS/United States
- P01-CA-65493/CA/NCI NIH HHS/United States
- P01 CA111412/CA/NCI NIH HHS/United States
- R01-HL-55417/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials