DNA demethylation in the Arabidopsis genome - PubMed (original) (raw)
DNA demethylation in the Arabidopsis genome
Jon Penterman et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Cytosine DNA methylation is considered to be a stable epigenetic mark, but active demethylation has been observed in both plants and animals. In Arabidopsis thaliana, DNA glycosylases of the DEMETER (DME) family remove methylcytosines from DNA. Demethylation by DME is necessary for genomic imprinting, and demethylation by a related protein, REPRESSOR OF SILENCING1, prevents gene silencing in a transgenic background. However, the extent and function of demethylation by DEMETER-LIKE (DML) proteins in WT plants is not known. Using genome-tiling microarrays, we mapped DNA methylation in mutant and WT plants and identified 179 loci actively demethylated by DML enzymes. Mutations in DML genes lead to locus-specific DNA hypermethylation. Reintroducing WT DML genes restores most loci to the normal pattern of methylation, although at some loci, hypermethylated epialleles persist. Of loci demethylated by DML enzymes, >80% are near or overlap genes. Genic demethylation by DML enzymes primarily occurs at the 5' and 3' ends, a pattern opposite to the overall distribution of WT DNA methylation. Our results show that demethylation by DML DNA glycosylases edits the patterns of DNA methylation within the Arabidopsis genome to protect genes from potentially deleterious methylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
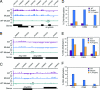
Mapping genome-wide DNA methylation in WT and ros1–3; dml2–1; dml3–1 identifies DML target loci. (A) Gene diagrams of the DME family members. Boxed regions are exons, and lines are introns. Blue exons encode the helix–hairpin–helix DNA glycosylase domain, and pink and orange exons encode conserved domains of unknown function (12). Black exons encode amino acids not shared between DML proteins. The position of ros1–3, ros1–4, ros1–5, dml2–1, and dml3–1 T-DNA insertions is marked by a triangle. (B) Scatter plot showing the correlation between WT and ros1–3; dml2–1; dml3–1 microarray experiments. The correlation coefficient (r) of the two data sets is 0.97. (C) Example of tiling microarray data. The top scale is the position in base pairs on chromosome 1. Each bar represents a single probe log2 signal ratio (5′-methylcytosine antibody pull-down/input) for WT and mutant data sets. For the [WT–mutant] data set, each bar represents the subtraction of a mutant log2 signal ratio from the corresponding WT log2 signal ratio, and a negative value in the [WT–mutant] data set is indicative of mutant hypermethylation. Genes are represented by black boxes; ones above the line are oriented 5′ to 3′ from left to right, and ones below are oriented 5′ to 3′ from right to left. An arrow indicates a locus hypermethylated in the mutant. Notice how the methylation profile flanking this locus is relatively similar between WT and mutant. (D) Genomic location of DML target loci. Shown are chromosomes 1–5 and their centromeres (red circles). To the right of each chromosome are horizontal lines that indicate the positions of each hypermethylated locus in the ros1–3; dml2–1; dml3–1 genome.
Fig. 2.
Bisulfite sequencing confirms methylation profiles of WT and mutant. (A–C) Examples of tiling microarray data showing three loci that were confirmed to be hypermethylated in ros1–3; dml2–1; dml3–1. The black bars above the [WT–mutant] data set indicate the positions of bisulfite sequencing. (D–F) Bisulfite sequencing data showing percents of CG, CNG, and CNN methylation for loci shown in A, B, and C, respectively, in WT, mutant, and their F1 progeny. See
SI Tables 4 and 5
for more details.
Fig. 3.
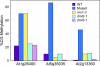
Bisulfite sequencing of single mutants shows that loci are demethylated exclusively by a single DML or redundantly by multiple DMLs. Graphed are the CG methylation levels of At1g26400, At5g35935, and At2g13360 in WT, triple mutant, ros1–3, dml2–1, and dml3–1 backgrounds. For all loci, the behavior of CG methylation is representative of methylation at CNG and CNN sites.
Fig. 4.
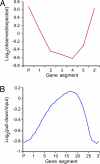
Distribution of DNA methylation within genes hypermethylated in ros1–3; dml2–1; dml3–1 (A) and within genes in WT (B). (A) Eighty-five genes hypermethylated in ros1–3; dml2–1; dml3–1 were divided into five equal size segments plus a 500-bp 5′ and 3′ segment. Plotted is the log2 ratio of observed hypermethylated segments over the number expected through chance alone. The distribution is significantly different than expected (χ2 = 24.2; df = 6; P < 0.001). (B) Plotted is the average log2 signal ratio (pull-down/input) for each segment from 21,583 Arabidopsis genes. Genes were divided into 25 equal size segments plus five 100-bp segments on both the 5′ and 3′ ends.
Similar articles
- DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases.
Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marín MI, Martínez-Macías MI, Ariza RR, Roldán-Arjona T. Morales-Ruiz T, et al. Proc Natl Acad Sci U S A. 2006 May 2;103(18):6853-8. doi: 10.1073/pnas.0601109103. Epub 2006 Apr 19. Proc Natl Acad Sci U S A. 2006. PMID: 16624880 Free PMC article. - DNA demethylation: a lesson from the garden.
Ikeda Y, Kinoshita T. Ikeda Y, et al. Chromosoma. 2009 Feb;118(1):37-41. doi: 10.1007/s00412-008-0183-3. Epub 2008 Oct 7. Chromosoma. 2009. PMID: 18839198 Review. - Genome-wide demethylation of Arabidopsis endosperm.
Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D. Hsieh TF, et al. Science. 2009 Jun 12;324(5933):1451-4. doi: 10.1126/science.1172417. Science. 2009. PMID: 19520962 Free PMC article. - Control of DEMETER DNA demethylase gene transcription in male and female gamete companion cells in Arabidopsis thaliana.
Park JS, Frost JM, Park K, Ohr H, Park GT, Kim S, Eom H, Lee I, Brooks JS, Fischer RL, Choi Y. Park JS, et al. Proc Natl Acad Sci U S A. 2017 Feb 21;114(8):2078-2083. doi: 10.1073/pnas.1620592114. Epub 2017 Jan 27. Proc Natl Acad Sci U S A. 2017. PMID: 28130550 Free PMC article. - Genome demethylation and imprinting in the endosperm.
Bauer MJ, Fischer RL. Bauer MJ, et al. Curr Opin Plant Biol. 2011 Apr;14(2):162-7. doi: 10.1016/j.pbi.2011.02.006. Epub 2011 Mar 23. Curr Opin Plant Biol. 2011. PMID: 21435940 Free PMC article. Review.
Cited by
- Retrotransposon-driven environmental regulation of FLC leads to adaptive response to herbicide.
Raingeval M, Leduque B, Baduel P, Edera A, Roux F, Colot V, Quadrana L. Raingeval M, et al. Nat Plants. 2024 Nov;10(11):1672-1681. doi: 10.1038/s41477-024-01807-8. Epub 2024 Sep 27. Nat Plants. 2024. PMID: 39333353 - Control of DNA demethylation by superoxide anion in plant stem cells.
Wang S, Liu M, Hu D, Dong Z, Zhao Z. Wang S, et al. Nat Chem Biol. 2024 Sep 12. doi: 10.1038/s41589-024-01737-8. Online ahead of print. Nat Chem Biol. 2024. PMID: 39266722 - Transgenerational epigenetic inheritance during plant evolution and breeding.
Cao S, Chen ZJ. Cao S, et al. Trends Plant Sci. 2024 Nov;29(11):1203-1223. doi: 10.1016/j.tplants.2024.04.007. Epub 2024 May 28. Trends Plant Sci. 2024. PMID: 38806375 Review. - A DNA demethylase reduces seed size by decreasing the DNA methylation of AT-rich transposable elements in soybean.
Wang W, Zhang T, Liu C, Liu C, Jiang Z, Zhang Z, Ali S, Li Z, Wang J, Sun S, Chen Q, Zhang Q, Xie L. Wang W, et al. Commun Biol. 2024 May 21;7(1):613. doi: 10.1038/s42003-024-06306-2. Commun Biol. 2024. PMID: 38773248 Free PMC article. - Insights into plant regeneration: cellular pathways and DNA methylation dynamics.
Lee S, Park YS, Rhee JH, Chu H, Frost JM, Choi Y. Lee S, et al. Plant Cell Rep. 2024 Apr 18;43(5):120. doi: 10.1007/s00299-024-03216-9. Plant Cell Rep. 2024. PMID: 38634973 Free PMC article. Review.
References
- Chan SW, Henderson IR, Jacobsen SE. Nat Rev Genet. 2005;6:351–360. - PubMed
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. Nature. 2004;430:471–476. - PubMed
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Nat Genet. 2007;39:61–69. - PubMed
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Cell. 2006;126:1189–1201. - PubMed
- Tran RK, Henikoff JG, Zilberman D, Ditt RF, Jacobsen SE, Henikoff S. Curr Biol. 2005;15:154–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials