The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3'UTR-mediated repression of the let-7 microRNA target gene, hbl-1 - PubMed (original) (raw)
The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3'UTR-mediated repression of the let-7 microRNA target gene, hbl-1
Mona J Nolde et al. Dev Biol. 2007.
Abstract
The Puf family of RNA-binding proteins directs cell fates by regulating gene expression at the level of translation and RNA stability. Here, we report that the Caenorhabditis elegans pumilio homolog, puf-9, controls the differentiation of epidermal stem cells at the larval-to-adult transition. Genetic analysis reveals that loss-of-function mutations in puf-9 enhance the lethality and heterochronic phenotypes caused by mutations in the let-7 microRNA (miRNA), while suppressing the heterochronic phenotypes of lin-41, a let-7 target and homolog of Drosophila Brat. puf-9 interacts with another known temporal regulator hbl-1, the Caenorhabditis elegans ortholog of hunchback. We present evidence demonstrating that puf-9 is required for the 3'UTR-mediated regulation of hbl-1, in both the hypodermis and the ventral nerve cord. Finally, we show that this regulation is dependent on a region of the hbl-1 3'UTR that contains putative Puf family binding sites as well as binding sites for the let-7 miRNA family, suggesting that puf-9 and let-7 may mediate hypodermal seam cell differentiation by regulating common targets.
Figures
Figure 1
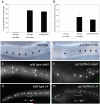
puf-9 depletion enhances the lethal bursting and heterochronic phenotypes of let-7(ts) mutants. (A) let-7(n2853ts) animals exposed to puf-9(RNAi) show an increase in the number of animals that die due to bursting at the adult stage (n=533) compared to let-7(n2853ts) animals grown on a control mock RNAi (n=1114). puf-9(ok1136) mutants (n=746) and wild type animals on puf-9(RNAi) (n=483) showed similar survival rates as let-7(n2853ts) animals on mock RNAi. 100% survival rate was seen in wild type animals grown on mock RNAi (n=652). (B) A cartoon of the intron/exon structure of the puf-9 gene (contained on the W06B11.2 cosmid). The relative position of the region deleted in the puf-9(ok1136) allele is boxed and shaded. The Pum-HD region is indicated within the ok1136 deletion region. The puf-9(ok1136) deletion removes 1581 nt beginning 154 nt into exon 4 and ending 77 nt into exon 8 (nt 5727 to 7309 in the W06B11.2 cosmid sequence). This deletion ends 632 nt upstream of the predicted stop codon and causes a frame-shift to create a premature stop codon 29 nt downstream of the deletion break. (C-E) Depletion of let-7 and puf-9 gene products results in a similar vulval bursting phenotype. Nomarski DIC images of a wild type adult (C, vulva is marked with an arrow), an adult stage let-7(n2853ts) animal with a burst vulva (D), and a puf-9(ok1136) adult animal showing a bursting phenotype similar to let-7(lf) mutants (E). (F) puf-9(RNAi) enhances the lack of adult alae phenotype of let-7(lf) mutants. When grown on puf-9(RNAi), less than half of the let-7(n2853ts) animals displayed any alae at the young adult stage (n=40) compared to wild type animals exposed to puf-9(RNAi) (n=27) or let-7(n2853ts) mutants on mock RNAi (n=61).
Figure 2
puf-9 deletion mutants display abnormal alae and seam cell fusion phenotypes. (A) puf-9(ok1136) mutants (n=88) and puf_-9(RNAi)_ animals (n=157) show incomplete alae at the adult stage (89% and 94%, respectively) compared to 1% in wild type animals (n=129) (C,D) Nomarski DIC images of adult stage alae in a wild type animal (C, alae marked with arrows) and an adult stage puf-9(ok1136) animal showing incomplete alae (D, alae marked with arrows and the alae break marked with an arrow head). (B, E-H) Seam cell analysis of wIs79 animals carrying a gfp reporter marking the seam cell nuclei and adherens junctions. (B) Approximately half of the wIs79 animals exposed to puf-9(RNAi) (n=96) or in a puf-9(ok1136) mutant background (n=21) showed incomplete seam cell fusion at the young adult stage compared to wIs79 on mock RNAi (n=63). (E-H) GFP images of wIs79;puf-9(RNAi) animals. Fusion defects included gaps along the length of the seam cell boundary syncytium (F, gap edges marked by arrow heads) and single seam cell nuclei separated from the rest of the syncytium (F, the anterior boundary of a single unfused seam cell is marked by the arrow), indicating that a number of seam cells failed to properly fuse to the seam cell syncytium. puf-9(RNAi) animals also show seam cell nuclei that are abnormally spaced (F, set of nuclei between arrow heads and H, nuclei within bracket) and are often spatially displaced relative to one another (H, bracketed nuclei). (E) Wild type adult seam cell fusion (seam cell syncytium between arrows) and (G) anterior-posterior seam cell nuclei orientation (a seam cell pair is in the bracket). Anterior is to the left and ventral down in all images.
Figure 3
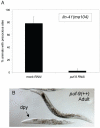
puf-9(RNAi) by feeding suppresses lin-41(lf) precocious alae. (A) lin-41(ma104) animals exposed to puf-9(RNAi) (n=35) showed an almost complete reduction in the number of animals that displayed alae in the early L4 stage compared to animals on mock RNAi (n=21). (B) Full length puf-9::gfp over-expressing animals showed dumpy (Dpy) phenotypes reminiscent of lin-41 and hbl-1 loss-of-function mutants. The transgenic animal in (B) is indicated by an arrow.
Figure 4
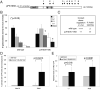
hbl-1(RNAi) suppresses puf-9(ok1136) adult alae phenotypes while puf-9(RNAi) is unable to suppress a strong loss-of-function hbl-1 allele. (A) hbl-1(RNAi) suppresses the puf-9(ok1136) adult alae phenotypes. The number of puf-9(ok1136) animals displaying incomplete alae was decreased on hbl-1(RNAi) (n=55) compared to puf-9(ok1136) mutants alone (n=59). (B, left data sets) puf-9 is unable to suppress the hbl-1(ve18) precocious alae phenotype. hbl-1(ve18) mutants on mock RNAi (n=76) show a complete penetrance of precocious alae in the early L4 stage and nearly all hbl-1(ve18); puf-9(RNAi) animals (n=96) continued to display this precocious alae defect. (B, middle and right data sets) The precocious alae phenotypes of a weak hbl-1 allele and hbl-1(RNAi) are partially suppressed by removal of puf-9. hbl-1(mg285) animals exposed to puf-9(RNAi) (n=133) show a reduction in the number of animals displaying precocious alae at the early L4 stage, compared with mock RNAi (n=77). Similarly, puf-9(ok1136) mutants on hbl-1(RNAi) (n=75) suppressed the precocious alae seen with hbl-1(RNAi) exposure in a wild type background (n=69). puf-9(ok1136) mutant animals did not show any precocious alae on their own when fed mock RNAi (n=43). Error bars indicate standard deviations.
Figure 5
A wild type copy of puf-9 is required for hbl-1 3’UTR reporter down-regulation in the hypodermis and the neurons. (A) A diagram of the heterologous reporter gene (pFS1038) used to assay hbl-1 3’UTR-directed regulation in the hypodermis, consisting of the wild type hbl-1 3’UTR fused downstream of the Escherichia coli lacZ gene and driven by the hypodermally expressed col-10 promoter (not to scale). (B, C) puf-9(ok1136) mutant animals carrying the pFS1038 reporter showed maintenance of β-galactosidase activity in the adult stage (17%), while the same construct in wild type animals showed expected levels of reporter gene down-regulation (2%) at the adult stage. Three independent staining trials were performed. The number of lines scored for each strain are, wild type: 1 line and puf-9(ok1136): 5 lines, n > 270 for all stages scored. For statistical analysis, an unpaired Student’s t test was performed between corresponding stages for the wild type and puf-9(ok1136) backgrounds. Error bars indicate standard deviations. (C) The correct down-regulation for each construct was compared to wild type and assigned values as follows: (++++) 0%−5% of animals with hypodermal expression at the adult stage; (+++) 5%−10%; (++) 10%−20%; (+) 20%−60%; (−) >60%. (D, E) A wild type copy of puf-9 is required for adult stage gfp::hbl-1 3’UTR reporter down-regulation in the VNC. (D) Late L4 (n=18) and adult stage (n=67) puf-9(ok1136) mutants showed an increase in the number of animals with bright HBL-1/GFP in the VNC compared to the same stages in wild type animals (L4, n=21 and adult, n=63). Wild type and puf-9(ok1136) animals were scored in three independent trials for adult GFP expression, and one trial for L4 GFP expression. (E) Quantification of wild type and puf-9(ok1136) GFP levels in the VNC was performed for a sample of L1 and adult stage animals. puf-9(ok1136) mutants showed a significant up-regulation of GFP at the L1 (n=8) and adult stages (n=12) compared to wild type animals (L1, n=4 and adult, n=10). The levels of GFP expression were calculated by normalizing the quantity of average GFP signal in puf-9(ok1136) mutants against the average GFP signal in wild type animals (for L1 and adult stages). Statistics were performed using an unpaired Student’s t test.
Figure 6
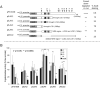
hbl-1 is regulated by multiple sequences, including the NRE region, in its 3’UTR. (A) Schematic of the hbl-1 3’UTR deletion constructs used to determine the necessary regulatory region of the hbl-1 3’UTR. Each construct shares a common col-10 promoter and contains the indicated portions of the Caenorhabditis elegans hbl-1 3’UTR fused to the Escherichia coli lacZ reporter gene. LCS are designated by arrowheads, with LCS 1 and LCS 2 specified by the double lines. LCE are shown as arrows, with LCE 1 designated as a dashed line. NRE1 and NRE2 are shown as black, numbered boxes within the construct. The values for correct down-regulation were assigned as in Figure 5. pFS1038 is the full-length reporter construct and contains the wild type 1402 bp hbl-1 3’UTR. (A, B) The deletion constructs all showed statistically significant up-regulation at the adult stage compared to wild type (students t test). The number of lines scored for each construct in (B) are pFS1038: 1 line (4 trials), pSJA2: 3 lines, pSJA3: 4 lines, pSJA4: 2 lines, pSJA5: 3 lines, and pMJC2: 4 lines, n > 50 for all stages scored. Error bars indicate standard deviations.
Fig. 7
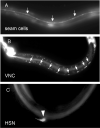
puf-9::gfp is expressed in the hypodermis and a subset of the neurons. Animals carrying a puf-9::gfp transcriptional fusion (MJC20) showed larval GFP expression in the lateral hypodermal seam cells (A, arrows) and non-seam cell hypodermis (A, diffuse staining). Additionally, GFP expression was noted in various neural cells including the ventral neural cord (VNC) (B, cell bodies marked by arrows), and the hermaphrodite specific neurons (HSN) (C, marked by an arrowhead).
Similar articles
- The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target.
Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. Lin SY, et al. Dev Cell. 2003 May;4(5):639-50. doi: 10.1016/s1534-5807(03)00124-2. Dev Cell. 2003. PMID: 12737800 - The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs.
Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. Abrahante JE, et al. Dev Cell. 2003 May;4(5):625-37. doi: 10.1016/s1534-5807(03)00127-8. Dev Cell. 2003. PMID: 12737799 - Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression.
Banerjee D, Slack F. Banerjee D, et al. Bioessays. 2002 Feb;24(2):119-29. doi: 10.1002/bies.10046. Bioessays. 2002. PMID: 11835276 Review. - The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function?
Spassov DS, Jurecic R. Spassov DS, et al. IUBMB Life. 2003 Jul;55(7):359-66. doi: 10.1080/15216540310001603093. IUBMB Life. 2003. PMID: 14584586 Review.
Cited by
- Alternative Polyadenylation in Triple-Negative Breast Tumors Allows NRAS and c-JUN to Bypass PUMILIO Posttranscriptional Regulation.
Miles WO, Lembo A, Volorio A, Brachtel E, Tian B, Sgroi D, Provero P, Dyson N. Miles WO, et al. Cancer Res. 2016 Dec 15;76(24):7231-7241. doi: 10.1158/0008-5472.CAN-16-0844. Epub 2016 Oct 10. Cancer Res. 2016. PMID: 27758885 Free PMC article. - The Puf-family RNA-binding protein PfPuf2 regulates sexual development and sex differentiation in the malaria parasite Plasmodium falciparum.
Miao J, Li J, Fan Q, Li X, Li X, Cui L. Miao J, et al. J Cell Sci. 2010 Apr 1;123(Pt 7):1039-49. doi: 10.1242/jcs.059824. Epub 2010 Mar 2. J Cell Sci. 2010. PMID: 20197405 Free PMC article. - Molecular architecture of a miRNA-regulated 3' UTR.
Didiano D, Hobert O. Didiano D, et al. RNA. 2008 Jul;14(7):1297-317. doi: 10.1261/rna.1082708. Epub 2008 May 7. RNA. 2008. PMID: 18463285 Free PMC article. - A microRNA network regulates proliferative timing and extracellular matrix synthesis during cellular quiescence in fibroblasts.
Suh EJ, Remillard MY, Legesse-Miller A, Johnson EL, Lemons JM, Chapman TR, Forman JJ, Kojima M, Silberman ES, Coller HA. Suh EJ, et al. Genome Biol. 2012 Dec 22;13(12):R121. doi: 10.1186/gb-2012-13-12-r121. Genome Biol. 2012. PMID: 23259597 Free PMC article. - Nanos genes and their role in development and beyond.
De Keuckelaere E, Hulpiau P, Saeys Y, Berx G, van Roy F. De Keuckelaere E, et al. Cell Mol Life Sci. 2018 Jun;75(11):1929-1946. doi: 10.1007/s00018-018-2766-3. Epub 2018 Feb 3. Cell Mol Life Sci. 2018. PMID: 29397397 Free PMC article. Review.
References
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. - PubMed
- Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57:49–57. - PubMed
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. - PubMed
- Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat Cell Biol. 1999;1:431–437. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM062594-05/GM/NIGMS NIH HHS/United States
- GM64701/GM/NIGMS NIH HHS/United States
- R01 GM064701/GM/NIGMS NIH HHS/United States
- GM62594/GM/NIGMS NIH HHS/United States
- R01 GM062594/GM/NIGMS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 GM064701-04/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials