A single IGF1 allele is a major determinant of small size in dogs - PubMed (original) (raw)
. 2007 Apr 6;316(5821):112-5.
doi: 10.1126/science.1137045.
Carlos D Bustamante, Kevin Chase, Melissa M Gray, Keyan Zhao, Lan Zhu, Badri Padhukasahasram, Eric Karlins, Sean Davis, Paul G Jones, Pascale Quignon, Gary S Johnson, Heidi G Parker, Neale Fretwell, Dana S Mosher, Dennis F Lawler, Ebenezer Satyaraj, Magnus Nordborg, K Gordon Lark, Robert K Wayne, Elaine A Ostrander
Affiliations
- PMID: 17412960
- PMCID: PMC2789551
- DOI: 10.1126/science.1137045
A single IGF1 allele is a major determinant of small size in dogs
Nathan B Sutter et al. Science. 2007.
Erratum in
- Science. 2007 Jun 1;316(5829):1284
Abstract
The domestic dog exhibits greater diversity in body size than any other terrestrial vertebrate. We used a strategy that exploits the breed structure of dogs to investigate the genetic basis of size. First, through a genome-wide scan, we identified a major quantitative trait locus (QTL) on chromosome 15 influencing size variation within a single breed. Second, we examined genetic variation in the 15-megabase interval surrounding the QTL in small and giant breeds and found marked evidence for a selective sweep spanning a single gene (IGF1), encoding insulin-like growth factor 1. A single IGF1 single-nucleotide polymorphism haplotype is common to all small breeds and nearly absent from giant breeds, suggesting that the same causal sequence variant is a major contributor to body size in all small dogs.
Figures
Fig. 1
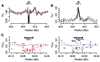
Relationships of skeletal size, SNP markers, IGF1 haplotype, and serum levels of the IGF1 protein in PWDs. (A) A mixed-model test for association between size and genotype. The association of three genotype categories (A1A1, A1A2, and A2A2) with skeletal size measurements was calculated with the use of all pairwise coefficients of consanguinity for 376 dogs. Each point represents a single SNP position on canine chromosome 15 and negative log P value for the association statistic. (B) PWD IGF1 haplotypes and mean skeletal size. Haplotypes were inferred for 20 markers spanning the IGF1 gene (chromosome 15: 44,212,792 to 44,278,140, CanFam1). Out of the 720 chromosomes with successful inference, 96% carry one of just two haplotypes, B and I, identical to haplotypes inferred for small and giant dogs, respectively (Fig. 3). Data are graphed as a histogram for each genotype: B/B (closed triangle, black line), B/I (open square, dashed line), and I/I (closed circle, gray line). (C) Serum levels of IGF1 protein (ng/ml) as a function of haplotype. Serum levels of IGF1 protein were assayed in 31 PWDs carrying haplotypes B and I. Box plots show the median (center line in box), first and third quartile (box ends), and maximum and minimum values (whiskers) obtained for each category: homozygous B/B (n = 15), heterozygous B/I (n = 7), and homozygous I/I (n = 9).
Fig. 2
Signatures of recent selection on the IGF1 locus across 22 small and giant dog breeds. (A) Heterozygosity ratio (_H_R) for small versus giant dogs. (B) Genetic differentiation (_F_ST) for small versus giant dogs. For both (A) and (B), a sliding 10-SNP window across IGF1 was used. Dashed lines delimit the 95% confidence intervals based on nonparametric bootstrap resampling. The IGF1 gene interval is indicated above the graphs as a red box drawn to scale. (C) Observed heterozygosity (_H_Obs) of SNPs near IGF1 typed in small breeds (<9 kg) and giant breeds (>30 kg). Small breeds have a reduction in observed heterozygosity compared with that of giant breeds. Red and blue points are average observed heterozygosity in small and giant breeds, respectively. Dashed lines are locally weighted scatterplot smoothing (LOWESS) best fit to the data. The IGF1 gene is shown as a black bar with exons indicated by vertical lines.
Fig. 3
Evidence of association and IGF1 haplotypes for 14 small and 9 giant breeds. (A) Mann-Whitney U (MWU) P values for tests of association between individual SNPs and body size (small versus giant) for 116 SNPs on chromosome 15 and 83 SNPs on five control chromosomes. The dashed line indicates Bonferroni correction for multiple tests. Only breeds with data for at least 10 chromosomes were included (14 small and 9 giant breeds). (B) Haplotypes for the 20 markers spanning the small breed sweep interval near IGF1. The haplotypes were inferred independently in each breed. For each individual, fractional chromosome counts were summed for all haplotypes with at least 5% probability according to the haplotype inference software program PHASE. Chromosome sums for each breed were rounded to integer values; several breeds have odd numbers of chromosomes due to rounding error. Only inferred haplotypes carried by at least three dog chromosomes total (i.e., >0.5% frequency overall) are shown. Sequence reads collected from golden jackal (Canis aureus) were used to determine the ancestral allele for each SNP. The haplotypes are rows labeled A to L, and marker alleles are colored yellow for ancestral state (matching the nucleotide observed in the golden jackal) and blue for derived state. SNP positions within IGF1 are shown at the top with IGF1 introns (horizontal line) and exons (vertical bars) indicated. (C) Breed name and the average size of adult males in kilograms are provided. Small breeds less than 9 kg and giant breeds greater than 30 kg are grouped for totals shown at the far right.
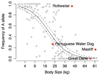
Fig. 4
Association of body size and frequency of the SNP 5 A allele. Binomial regression of allele frequency on square root of mean breed mass. Dashed lines indicate the 95% confidence interval on the predicted equation line as estimated from nonparametric bootstrap resampling. Between 5 and 109 (median = 22) dogs were genotyped for each of 143 breeds. The PWD is highlighted in red along with three giant breeds that have larger breed average masses than is predicted by their SNP 5 allele frequency.
Similar articles
- Derived variants at six genes explain nearly half of size reduction in dog breeds.
Rimbault M, Beale HC, Schoenebeck JJ, Hoopes BC, Allen JJ, Kilroy-Glynn P, Wayne RK, Sutter NB, Ostrander EA. Rimbault M, et al. Genome Res. 2013 Dec;23(12):1985-95. doi: 10.1101/gr.157339.113. Epub 2013 Sep 11. Genome Res. 2013. PMID: 24026177 Free PMC article. - Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping.
Vaysse A, Ratnakumar A, Derrien T, Axelsson E, Rosengren Pielberg G, Sigurdsson S, Fall T, Seppälä EH, Hansen MS, Lawley CT, Karlsson EK; LUPA Consortium; Bannasch D, Vilà C, Lohi H, Galibert F, Fredholm M, Häggström J, Hedhammar A, André C, Lindblad-Toh K, Hitte C, Webster MT. Vaysse A, et al. PLoS Genet. 2011 Oct;7(10):e1002316. doi: 10.1371/journal.pgen.1002316. Epub 2011 Oct 13. PLoS Genet. 2011. PMID: 22022279 Free PMC article. - The IGF1 small dog haplotype is derived from Middle Eastern grey wolves.
Gray MM, Sutter NB, Ostrander EA, Wayne RK. Gray MM, et al. BMC Biol. 2010 Feb 24;8:16. doi: 10.1186/1741-7007-8-16. BMC Biol. 2010. PMID: 20181231 Free PMC article. - Evolutionary genomics of dog domestication.
Wayne RK, vonHoldt BM. Wayne RK, et al. Mamm Genome. 2012 Feb;23(1-2):3-18. doi: 10.1007/s00335-011-9386-7. Epub 2012 Jan 22. Mamm Genome. 2012. PMID: 22270221 Review. - So many doggone traits: mapping genetics of multiple phenotypes in the domestic dog.
Rimbault M, Ostrander EA. Rimbault M, et al. Hum Mol Genet. 2012 Oct 15;21(R1):R52-7. doi: 10.1093/hmg/dds323. Epub 2012 Aug 9. Hum Mol Genet. 2012. PMID: 22878052 Free PMC article. Review.
Cited by
- Identification of functional rare coding variants in IGF-1 gene in humans with exceptional longevity.
Ali A, Zhang Z, Gao T, Aleksic S, Gavathiotis E, Barzilai N, Milman S. Ali A, et al. bioRxiv [Preprint]. 2024 Oct 13:2024.10.11.617885. doi: 10.1101/2024.10.11.617885. bioRxiv. 2024. PMID: 39416202 Free PMC article. Preprint. - The resistance of domestic canine skin-derived fibroblasts to oxidative and non-oxidative chemical injury: implications of breed and body size.
Harper JM, Hicks M, Jiménez AG. Harper JM, et al. Geroscience. 2024 Sep 24. doi: 10.1007/s11357-024-01358-y. Online ahead of print. Geroscience. 2024. PMID: 39316259 - Impact of ginger powder (Zingiber officinale) supplementation on the performance, biochemical parameters, antioxidant status, and rumen fermentation in Ossimi rams.
Ali ME, Alsalamah SA, Al-Thubyani SA, Baazaoui N, Ahmed AE, Nasser MA, Nasr HA. Ali ME, et al. Vet World. 2024 Jul;17(7):1619-1628. doi: 10.14202/vetworld.2024.1619-1628. Epub 2024 Jul 26. Vet World. 2024. PMID: 39185052 Free PMC article. - Signatures of selection in Angus and Hanwoo beef cattle using imputed whole genome sequence data.
Nawaz MY, Savegnago RP, Lim D, Lee SH, Gondro C. Nawaz MY, et al. Front Genet. 2024 Aug 2;15:1368710. doi: 10.3389/fgene.2024.1368710. eCollection 2024. Front Genet. 2024. PMID: 39161420 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM063056/GM/NIGMS NIH HHS/United States
- 5T32 HG002536/HG/NHGRI NIH HHS/United States
- R01 GM063056-06/GM/NIGMS NIH HHS/United States
- 063056/PHS HHS/United States
- P50 HG002790/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous