Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis - PubMed (original) (raw)
Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis
Song Mao et al. Cell Microbiol. 2007 Aug.
Abstract
Porphyromonas gingivalis can inhibit chemically induced apoptosis in primary cultures of gingival epithelial cells through blocking activation of the effector caspase-3. The anti-apoptotic phenotype of P. gingivalis is conserved across strains and does not depend on the presence of fimbriae, as fimbriae-deficient mutants and a naturally occurring non-fimbriated strain were able to impede apoptosis. To dissect the survival pathways modulated by P. gingivalis, protein and gene expression of a number of components of apoptotic death pathways were investigated. P. gingivalis infection of epithelial cells resulted in the phosphorylation of JAK1 and Stat3. Quantitative real-time reverse transcription polymerase chain reaction showed that expression of Survivin and Stat3 itself, targets of activated Stat3, were elevated in P. gingivalis-infected cells. siRNA knockdown of JAK1, in combination with knockdown of Akt, abrogated the ability of P. gingivalis to block apoptosis. In contrast, cIAP-1 and cIAP-2 were not differentially regulated at either the protein or mRNA levels by P. gingivalis. One mechanism by which P. gingivalis can block apoptotic pathways in gingival epithelial cells therefore is through manipulation of the JAK/Stat pathway that controls the intrinsic mitochondrial cell death pathways. Induction of a pro-survival phenotype may prevent programmed host cell death and aid survival of P. gingivalis within gingival epithelial cells.
Figures
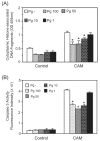
Fig. 1
Porphyromonas gingivalis infection protects gingival epithelial cells against camptothecin-induced apoptosis in a dose-dependent manner. Primary GECs were infected with P. gingivalis 33277 for 20 h at moi of 1, 10, 50 and 100 or were mock-infected (Pg-). Camptothecin (CAM) at 1 μgml−1 was added to the cells for an additional 4 h. Control represents GEC without CAM treatment. A. ELISA for histone-associated DNA fragments in the cytoplasm. B. Caspase-3 activity assessed by cleavage of the fluorescent substrate Ac-DEVD-AMC (arbitrary units). Error bars represent SD, n = 3. An asterisk denotes statistical difference of _P. gingivalis_-infected from mock-infected, P < 0.01, _t_-test.
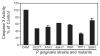
Fig. 2
The anti-apoptotic properties of P. gingivalis are conserved across strains and are not dependent on the presence of fimbriae. GECs were infected with P. gingivalis wild type and mutant strains at moi 50 for 20 h, or were mock-infected (Pg-). YPF1 is a FimA (long fimbriae) mutant, and SMF1 is a Mfa (short fimbriae) mutant. Camptothecin at 1 μgml−1 was added to the cells for an additional 4 h. Cells were lysed, and caspase-3 activity assessed by cleavage of the fluorescent substrate Ac-DEVD-AMC. Data are expressed as per cent fluorescent intensity (arbitrary units) of camptothecin alone control (CAM). Inhibition of caspase-3 was significant (P < 0.01, _t_-test) for all P. gingivalis strains.
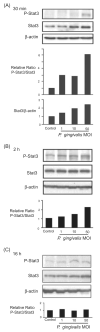
Fig. 3
Porphyromonas gingivalis infection transiently activates Stat3. GEC were infected with P. gingivalis at moi of 1, 10 and 50, or were mock-infected (control) for 30 min (A), 2 h (B) or 16 h (C). Cell lysates were analysed by immunoblotting with antibodies to Stat3, or phosphorylated (P-) Stat3. Blots were then stripped and probed with β-actin antibodies as a loading control. Blots were analysed by scanning densitometry and ratios of P-Stat3/Stat3 and Stat3/μ-actin determined relative to ratios in control cells. Data are representative of three independent experiments.
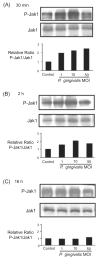
Fig. 4
Porphyromonas gingivalis infection transiently activates Jak1. GEC were infected with P. gingivalis at moi of 1, 10 and 50, or were mock-infected (control) for 30 min (A), 2 h (B) or 16 h (C). Cell lysates were analysed by immunoblotting with antibodies to Jak1, or phosphorylated (P-) Jak1. Blots were analysed by scanning densitometry and ratios of P-Jak1/Jak1 determined relative to ratios in control cells. Data are representative of three independent experiments.
Fig. 5
Simultaneous knockdown of Jak1 and Akt abrogates the anti-apoptotic activity of P. gingivalis. A. GECs were transfected with siRNA to Akt, Jak1, or both Akt and JAK. Controls were no target siRNA (SiNT) and transfection agent alone (control). GECs were infected with P. gingivalis at moi 50 for 20 h and then treated with camptothecin at 1 μgml−1 for 4 h. Caspase-3 activity was determined by a fluorescent substrate assay and data are expressed as per cent of mock-infected camptothecin treated cells (CAM). An asterisk denotes statistically significant (P < 0.01, _t_-test) from control and siNT. B. Scanning densiotometry of JAK1 (left panel) or Akt (right panel) expression in GECs after transfection with siRNA to Jak1, Akt or AKt and Jak1. Protein levels were determined by Western blotting and are expressed as per cent of the NT-siRNA control. Data are representative of three independent experiments.
Fig. 6
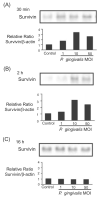
Porphyromonas gingivalis infection transiently increases levels of survivin. GEC were infected with P. gingivalis at moi of 1, 10 and 50, or were mock-infected (control) for 30 min (A), 2 h (B) or 16 h (C). Cell lysates were analysed by immunoblotting with antibodies to survivin. Blots were then stripped and probed with β-actin antibodies as a loading control (not shown). Blots were analysed by scanning densitometry and ratios of survivin/β-actin determined relative to ratios in control cells. Data are representative of three independent experiments.
Fig. 7
Porphyromonas gingivalis infection does not elevate levels of cIAP-1 or cIAP-2. GEC were infected with P. gingivalis at moi of 1, 10 and 50, or were mock-infected (control) for 30 min (A), 2 h (B) or 16 h (C). Cell lysates were analysed by immunoblotting with antibodies cIAP-1 or cIAP-2. Blots were then stripped and probed with β-actin antibodies as a loading control (not shown). Blots were analysed by scanning densitometry and ratios of cIAP/β-actin determined relative to ratios in control cells. Data are representative of three independent experiments.
Fig. 8
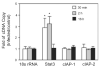
Stat3, but not cIAP-1/2, gene expression is transiently increased by P. gingivalis infection. Gene expression was measured by quantitative RT-PCR on mRNA from GEC infected with P. gingivalis at moi 50 for the times indicated, or mock-infected (control). 18sRNA was included as an internal control. Fold change was calculated by dividing the copy number of the gene transcript in P. gingivalis infected cells by the copy number in control cells. Significant differences infected/control (P < 0.01, _t_-test) are labelled with an asterisk.
Similar articles
- Porphyromonas gingivalis infection sequesters pro-apoptotic Bad through Akt in primary gingival epithelial cells.
Yao L, Jermanus C, Barbetta B, Choi C, Verbeke P, Ojcius DM, Yilmaz O. Yao L, et al. Mol Oral Microbiol. 2010 Apr;25(2):89-101. doi: 10.1111/j.2041-1014.2010.00569.x. Mol Oral Microbiol. 2010. PMID: 20331797 Free PMC article. - Porphyromonas gingivalis modulates Pseudomonas aeruginosa-induced apoptosis of respiratory epithelial cells through the STAT3 signaling pathway.
Li Q, Pan C, Teng D, Lin L, Kou Y, Haase EM, Scannapieco FA, Pan Y. Li Q, et al. Microbes Infect. 2014 Jan;16(1):17-27. doi: 10.1016/j.micinf.2013.10.006. Epub 2013 Oct 16. Microbes Infect. 2014. PMID: 24140557 - Invasion of Gingival Epithelial Cells by Porphyromonas gingivalis.
Takeuchi H, Amano A. Takeuchi H, et al. Methods Mol Biol. 2021;2210:215-224. doi: 10.1007/978-1-0716-0939-2_21. Methods Mol Biol. 2021. PMID: 32815142 - Dual lifestyle of Porphyromonas gingivalis in biofilm and gingival cells.
Sakanaka A, Takeuchi H, Kuboniwa M, Amano A. Sakanaka A, et al. Microb Pathog. 2016 May;94:42-7. doi: 10.1016/j.micpath.2015.10.003. Epub 2015 Oct 9. Microb Pathog. 2016. PMID: 26456558 Review.
Cited by
- Periodontitis: from microbial immune subversion to systemic inflammation.
Hajishengallis G. Hajishengallis G. Nat Rev Immunol. 2015 Jan;15(1):30-44. doi: 10.1038/nri3785. Nat Rev Immunol. 2015. PMID: 25534621 Free PMC article. Review. - The role of periodontitis in cancer development, with a focus on oral cancers.
Farhad SZ, Karbalaeihasanesfahani A, Dadgar E, Nasiri K, Esfahaniani M, Nabi Afjadi M. Farhad SZ, et al. Mol Biol Rep. 2024 Jul 15;51(1):814. doi: 10.1007/s11033-024-09737-6. Mol Biol Rep. 2024. PMID: 39008163 Review. - Emerging role of bacteria in oral carcinogenesis: a review with special reference to perio-pathogenic bacteria.
Perera M, Al-Hebshi NN, Speicher DJ, Perera I, Johnson NW. Perera M, et al. J Oral Microbiol. 2016 Sep 26;8:32762. doi: 10.3402/jom.v8.32762. eCollection 2016. J Oral Microbiol. 2016. PMID: 27677454 Free PMC article. Review. - Immunometabolic and potential tumor-promoting changes in 3D cervical cell models infected with bacterial vaginosis-associated bacteria.
Maarsingh JD, Łaniewski P, Herbst-Kralovetz MM. Maarsingh JD, et al. Commun Biol. 2022 Jul 22;5(1):725. doi: 10.1038/s42003-022-03681-6. Commun Biol. 2022. PMID: 35869172 Free PMC article. - Filifactor alocis interactions with gingival epithelial cells.
Moffatt CE, Whitmore SE, Griffen AL, Leys EJ, Lamont RJ. Moffatt CE, et al. Mol Oral Microbiol. 2011 Dec;26(6):365-73. doi: 10.1111/j.2041-1014.2011.00624.x. Epub 2011 Sep 13. Mol Oral Microbiol. 2011. PMID: 22053964 Free PMC article.
References
- Aaronson DS, Horvath CM. A road map for those who don’t know JAK-STAT. Science. 2002;296:1653–1655. - PubMed
- Albandar JM. Epidemiology and risk factors of periodontal diseases. Dent Clin North Am. 2005;49:517–532. - PubMed
- Belton CM, Izutsu KT, Goodwin PC, Park Y, Lamont RJ. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell Microbiol. 1999;1:215–223. - PubMed
- Beyaert R, Van Loo G, Heyninck K, Vandenabeele P. Signaling to gene activation and cell death by tumor necrosis factor receptors and Fas. Int Rev Cytol. 2002;214:225–272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DE011111/DE/NIDCR NIH HHS/United States
- R01 DE016593/DE/NIDCR NIH HHS/United States
- R37 DE011111/DE/NIDCR NIH HHS/United States
- DE11111/DE/NIDCR NIH HHS/United States
- R01 DE016593-02/DE/NIDCR NIH HHS/United States
- R01 DE016715/DE/NIDCR NIH HHS/United States
- DE16715/DE/NIDCR NIH HHS/United States
- R56 DE016593/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous