CpG methylation is targeted to transcription units in an invertebrate genome - PubMed (original) (raw)
CpG methylation is targeted to transcription units in an invertebrate genome
Miho M Suzuki et al. Genome Res. 2007 May.
Abstract
DNA is methylated at the dinucleotide CpG in genomes of a wide range of plants and animals. Among animals, variable patterns of genomic CpG methylation have been described, ranging from undetectable levels (e.g., in Caenorhabditis elegans) to high levels of global methylation in the vertebrates. The most frequent pattern in invertebrate animals, however, is mosaic methylation, comprising domains of methylated DNA interspersed with unmethylated domains. To understand the origin of mosaic DNA methylation patterns, we examined the distribution of DNA methylation in the Ciona intestinalis genome. Bisulfite sequencing and computational analysis revealed methylated domains with sharp boundaries that strongly colocalize with approximately 60% of transcription units. By contrast, promoters, intergenic DNA, and transposons are not preferentially targeted by DNA methylation. Methylated transcription units include evolutionarily conserved genes, whereas the most highly expressed genes preferentially belong to the unmethylated fraction. The results lend support to the hypothesis that CpG methylation functions to suppress spurious transcriptional initiation within infrequently transcribed genes.
Figures
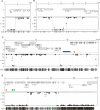
Figure 1.
A mosaic pattern of methlyated domains (MDs) and unmethylated domains (UMDs) in the C. intestinalis genome. Bisulfite sequencing showed the DNA methylation status at every CpG site in ribosomal protein S6 (scaffold 13: 227234–231211) (A), a randomly selected region of chromosome 9q (2421536–2429959) (B), a region of chromosome 7q corresponding to 29,300 bp of cosmid insert cos41 (C), and a region of chromosome 4q corresponding to 38,500 bp of cosmid insert cos2 (D). The top panels show gene annotation and plots of CpG methylation are shown below. Each dot represents a specific CpG corresponding to the CpG map at the bottom of each figure. MD/UMD boundaries are shown by broken lines. Repetitive sequences Cifo-1 (blue), Cimi-1 (green), and Cics-1 (orange) are shown.
Figure 2.
Methylated domains (MDs) within two 1-Mb genomic regions from chromosome 7q (A) and chromosome 4q (B) colocalize with transcription units. MDs (green bars) were inferred from CpG[o/e] ratios as described in the text. The inferred pattern was experimentally verified by methylation-sensitive PCR at sites marked by closed triangles (methylated) and open triangles (unmethylated). Yellow arrows show the size and orientation of predicted transcription units. The positions of bisulfite sequenced cosmid inserts cos41 and cos2 (see Fig. 1) are shown by black boxes. Green boxes indicate regions that were bisulfite sequenced in order to test the accuracy of predicted unmethylated domain (UMD)/MD boundaries (Supplemental Fig. 2). (C) Frequency of predicted genes that overlap to varying degrees with MDs. (D) Frequency of MDs that overlap to varying degrees with predicted genes. (E) Frequency of UMDs with gene overlap.
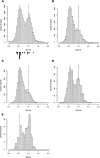
Figure 3.
Features that distinguish C. intestinalis methylated and unmethylated genes. (A) Histogram showing the frequency of all genes with CpG[o/e] frequencies between 0 and 2. CpG[o/e] of genes whose methylation status was analyzed by bisulfite sequencing are shown in closed (methylated) and open (unmethylated) triangles below. (B) Frequencies of genes that are evolutionarily conserved between C. intestinalis, Homo sapiens, and D. melanogaster plotted against CpG[o/e]. (C) Frequencies of C. intestinalis orthologs of human CpG island genes plotted against CpG[o/e]. (D) Frequencies of C. intestinalis orthologs of human non-CpG island genes plotted against CpG[o/e]. (E) Frequencies of 188 genes that recorded the highest number of EST hits (>500) in the UniGene database plotted against CpG[o/e].
Figure 4.
Variable DNA methylation of repetitive elements in C. intestinalis matches flanking DNA. Elements as named here or by Simmen and Bird (2000) are diagrammed as solid bars above each bisulfite profile showing primer pair location. A horizontal row within each box corresponds to one sequenced clone in which specific CpGs were methylated (solid) or unmethylated (open). Mutated CpG sites are shown as blanks within a row. (A) Results using primer sets within the element showed a mixture of methylated and unmethylated clones. (B) Locus-specific primers showed that the methylation patterns of two Cics-1 elements and two Cimi-1 elements at specific genomic locations are homogeneous and match the methylation status of flanking genomic DNA (open bars above). The locations of the elements analyzed in B are shown in Fig. 1.
Similar articles
- Evolutionary Transition of Promoter and Gene Body DNA Methylation across Invertebrate-Vertebrate Boundary.
Keller TE, Han P, Yi SV. Keller TE, et al. Mol Biol Evol. 2016 Apr;33(4):1019-28. doi: 10.1093/molbev/msv345. Epub 2015 Dec 29. Mol Biol Evol. 2016. PMID: 26715626 Free PMC article. - Nonmethylated transposable elements and methylated genes in a chordate genome.
Simmen MW, Leitgeb S, Charlton J, Jones SJ, Harris BR, Clark VH, Bird A. Simmen MW, et al. Science. 1999 Feb 19;283(5405):1164-7. doi: 10.1126/science.283.5405.1164. Science. 1999. PMID: 10024242 - Gradual transition from mosaic to global DNA methylation patterns during deuterostome evolution.
Okamura K, Matsumoto KA, Nakai K. Okamura K, et al. BMC Bioinformatics. 2010 Oct 15;11 Suppl 7(Suppl 7):S2. doi: 10.1186/1471-2105-11-S7-S2. BMC Bioinformatics. 2010. PMID: 21106124 Free PMC article. - The methyl-CpG binding domain and the evolving role of DNA methylation in animals.
Hendrich B, Tweedie S. Hendrich B, et al. Trends Genet. 2003 May;19(5):269-77. doi: 10.1016/S0168-9525(03)00080-5. Trends Genet. 2003. PMID: 12711219 Review. - Monitoring methylation changes in cancer.
Beier V, Mund C, Hoheisel JD. Beier V, et al. Adv Biochem Eng Biotechnol. 2007;104:1-11. doi: 10.1007/10_024. Adv Biochem Eng Biotechnol. 2007. PMID: 17290816 Review.
Cited by
- Is There a Relationship between DNA Methylation and Phenotypic Plasticity in Invertebrates?
Roberts SB, Gavery MR. Roberts SB, et al. Front Physiol. 2012 Jan 2;2:116. doi: 10.3389/fphys.2011.00116. eCollection 2012. Front Physiol. 2012. PMID: 22232607 Free PMC article. - Epigenetic machinery is functionally conserved in cephalopods.
Macchi F, Edsinger E, Sadler KC. Macchi F, et al. BMC Biol. 2022 Sep 14;20(1):202. doi: 10.1186/s12915-022-01404-1. BMC Biol. 2022. PMID: 36104784 Free PMC article. - Potential Role of DNA Methylation as a Driver of Plastic Responses to the Environment Across Cells, Organisms, and Populations.
Bogan SN, Yi SV. Bogan SN, et al. Genome Biol Evol. 2024 Feb 1;16(2):evae022. doi: 10.1093/gbe/evae022. Genome Biol Evol. 2024. PMID: 38324384 Free PMC article. - Control of genic DNA methylation in Arabidopsis.
Inagaki S, Kakutani T. Inagaki S, et al. J Plant Res. 2010 May;123(3):299-302. doi: 10.1007/s10265-010-0338-1. Epub 2010 Apr 3. J Plant Res. 2010. PMID: 20364290 - What drives phenotypic divergence among coral clonemates of Acropora palmata?
Durante MK, Baums IB, Williams DE, Vohsen S, Kemp DW. Durante MK, et al. Mol Ecol. 2019 Jul;28(13):3208-3224. doi: 10.1111/mec.15140. Epub 2019 Jul 7. Mol Ecol. 2019. PMID: 31282031 Free PMC article.
References
- Bird A.P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. - PubMed
- Bird A.P. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 1987;3:342–347.
- Bird A.P. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. - PubMed
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources