Analysis of PALB2/FANCN-associated breast cancer families - PubMed (original) (raw)
. 2007 Apr 17;104(16):6788-93.
doi: 10.1073/pnas.0701724104. Epub 2007 Apr 9.
Bing Xia, Nelly Sabbaghian, Jorge S Reis-Filho, Nancy Hamel, Guilan Li, Erik H van Beers, Lili Li, Tayma Khalil, Louise A Quenneville, Atilla Omeroglu, Aletta Poll, Pierre Lepage, Nora Wong, Petra M Nederlof, Alan Ashworth, Patricia N Tonin, Steven A Narod, David M Livingston, William D Foulkes
Affiliations
- PMID: 17420451
- PMCID: PMC1871863
- DOI: 10.1073/pnas.0701724104
Analysis of PALB2/FANCN-associated breast cancer families
Marc Tischkowitz et al. Proc Natl Acad Sci U S A. 2007.
Abstract
No more than approximately 30% of hereditary breast cancer has been accounted for by mutations in known genes. Most of these genes, such as BRCA1, BRCA2, TP53, CHEK2, ATM, and FANCJ/BRIP1, function in DNA repair, raising the possibility that germ line mutations in other genes that contribute to this process also predispose to breast cancer. Given its close relationship with BRCA2, PALB2 was sequenced in affected probands from 68 BRCA1/BRCA2-negative breast cancer families of Ashkenazi Jewish, French Canadian, or mixed ethnic descent. The average BRCAPRO score was 0.58. A truncating mutation (229delT) was identified in one family with a strong history of breast cancer (seven breast cancers in three female mutation carriers). This mutation and its associated breast cancers were characterized with another recently reported but unstudied mutation (2521delA) that is also associated with a strong family history of breast cancer. There was no loss of heterozygosity in tumors with either mutation. Moreover, comparative genomic hybridization analysis showed major similarities to that of BRCA2 tumors but with some notable differences, especially loss of 18q, a change that was previously unknown in BRCA2 tumors and less common in sporadic breast cancer. This study supports recent observations that PALB2 mutations are present, albeit not frequently, in breast cancer families. The apparently high penetrance noted in this study suggests that at least some PALB2 mutations are associated with a substantially increased risk for the disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
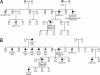
Fig. 1.
Pedigrees of the two studied families with PALB2 mutations. MM, malignant melanoma; PSU, primary site unknown; ∗, unconfirmed cancer diagnosis. (A) Pedigree A. (B) Pedigree B. In Family B, the mutation was not found in tumor samples from the proband's mother (BII.2), indicating that the mutation was inherited from the paternal side.
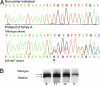
Fig. 2.
Sequence chromatogram showing WT exon 4 PALB2 sequence with the frameshift due to 229delT in the proband of Family A. The WT and mutant strand are indicated. The arrow points to where the T is deleted on the mutant strand. (A) LOH studies in PALB2 2521delA-related breast cancers. (B) A 115-bp region including the mutation _PALB2:2521delA was amplified by PCR from DNA extracted from patient III:2 (Fig. 1_B). Genotypes from this patient's blood (B) and macrodissected formalin-fixed tumor (T1, right breast tumor, diagnosed age 29; T2, left breast tumor, diagnosed age 46) and from an individual who does not carry the mutation (N) are shown. The larger WT band can be observed in all samples, whereas the fragment amplified from the mutated DNA strand (1 bp smaller) can be observed in the patient's blood and tumors but not in the noncarrier. There is no significant LOH of either the WT or mutant allele in the patient's tumors compared with the blood. A faint “shadow” band of the WT allele is present below the WT band in the mutation-negative control, and this band also appears at the same position as the mutant allele in B, T1, and T2. Tumors from AIII:1 and BIII:5 were also analyzed, and no LOH was detected (data not shown).
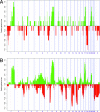
Fig. 3.
Frequency of DNA copy number changes in tumors arising in PALB2 and BRCA2 germline mutation carriers. (A) PALB2 cases [invasive breast cancer (n = 4) and lobular carcinoma in situ (n = 1)]. (B) BRCA2 tumors (n = 13). Individual BAC clones are plotted according to genomic location along the x axis. The proportion of tumors in which each clone is gained (green bars) or lost (red bars) is plotted along the y axis. Vertical dotted lines represent chromosome centromeres.
Fig. 4.
PALB2 protein structure and assessment of functional consequences of the 229delT and 2521delA mutations. (A) Schematic diagram of the protein showing predicted functional domains and the sites of the two truncating mutations studied. The extended reading frames after frameshifts are shown in orange. (B) 293T cells were transfected with the indicated plasmids, and N-terminally FLAG-HA-double tagged PALB2 proteins were precipitated with anti-FLAG M2 agarose beads. The abundance of tagged PALB2 proteins and BRCA2 in the precipitates was analyzed by Western blot. (C) DR-U2OS HR reporter cells were treated with control or PALB2 siRNAs and then cotransfected with pCBASce together with the pOZC or pOZC-PALB2 vectors, which contain seven silent base changes and are resistant to the siRNA used. GFP-positive cells were counted 72 h later. Data presented is from a representative experiment performed in duplicate. (D) Mitomycin C sensitivity of the EUFA1341 (FA-N;PALB2−/−) fibroblasts stably expressing the indicated PALB2 species. Results shown are the averages of three independent experiments, each performed in duplicate.
Similar articles
- Analysis of FANCB and FANCN/PALB2 fanconi anemia genes in BRCA1/2-negative Spanish breast cancer families.
García MJ, Fernández V, Osorio A, Barroso A, Llort G, Lázaro C, Blanco I, Caldés T, de la Hoya M, Ramón Y Cajal T, Alonso C, Tejada MI, San Román C, Robles-Díaz L, Urioste M, Benítez J. García MJ, et al. Breast Cancer Res Treat. 2009 Feb;113(3):545-51. doi: 10.1007/s10549-008-9945-0. Epub 2008 Feb 27. Breast Cancer Res Treat. 2009. PMID: 18302019 - Screening for BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish BRCA1/2-founder mutation-negative breast and/or ovarian cancer individuals.
Kuusisto KM, Bebel A, Vihinen M, Schleutker J, Sallinen SL. Kuusisto KM, et al. Breast Cancer Res. 2011 Feb 28;13(1):R20. doi: 10.1186/bcr2832. Breast Cancer Res. 2011. PMID: 21356067 Free PMC article. - Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer.
Casadei S, Norquist BM, Walsh T, Stray S, Mandell JB, Lee MK, Stamatoyannopoulos JA, King MC. Casadei S, et al. Cancer Res. 2011 Mar 15;71(6):2222-9. doi: 10.1158/0008-5472.CAN-10-3958. Epub 2011 Feb 1. Cancer Res. 2011. PMID: 21285249 Free PMC article. - PALB2/FANCN: recombining cancer and Fanconi anemia.
Tischkowitz M, Xia B. Tischkowitz M, et al. Cancer Res. 2010 Oct 1;70(19):7353-9. doi: 10.1158/0008-5472.CAN-10-1012. Epub 2010 Sep 21. Cancer Res. 2010. PMID: 20858716 Free PMC article. Review. - [The clinical importance of a genetic analysis of moderate-risk cancer susceptibility genes in breast and other cancer patients from the Czech Republic].
Pohlreich P, Kleibl Z, Kleiblová P, Janatová M, Soukupová J, Macháčková E, Házová J, Vašíčková P, Sťahlová Hrabincová E, Navrátilová M, Svoboda M, Foretová L. Pohlreich P, et al. Klin Onkol. 2012;25 Suppl:S59-66. Klin Onkol. 2012. PMID: 22920209 Review. Czech.
Cited by
- PALB2: a novel inactivating mutation in a Italian breast cancer family.
Balia C, Sensi E, Lombardi G, Roncella M, Bevilacqua G, Caligo MA. Balia C, et al. Fam Cancer. 2010 Dec;9(4):531-6. doi: 10.1007/s10689-010-9382-1. Fam Cancer. 2010. PMID: 20852946 - Evidence against PALB2 involvement in Icelandic breast cancer susceptibility.
Gunnarsson H, Arason A, Gillanders EM, Agnarsson BA, Johannesdottir G, Johannsson OT, Barkardottir RB. Gunnarsson H, et al. J Negat Results Biomed. 2008 Jul 17;7:5. doi: 10.1186/1477-5751-7-5. J Negat Results Biomed. 2008. PMID: 18637200 Free PMC article. - Analysis of large deletions in BRCA1, BRCA2 and PALB2 genes in Finnish breast and ovarian cancer families.
Pylkäs K, Erkko H, Nikkilä J, Sólyom S, Winqvist R. Pylkäs K, et al. BMC Cancer. 2008 May 26;8:146. doi: 10.1186/1471-2407-8-146. BMC Cancer. 2008. PMID: 18501021 Free PMC article. - Homologous Recombination and Replication Fork Protection: BRCA2 and More!
Feng W, Jasin M. Feng W, et al. Cold Spring Harb Symp Quant Biol. 2017;82:329-338. doi: 10.1101/sqb.2017.82.035006. Epub 2018 Apr 23. Cold Spring Harb Symp Quant Biol. 2017. PMID: 29686033 Free PMC article. - Mutation analysis of BRCA1, BRCA2, PALB2 and BRD7 in a hospital-based series of German patients with triple-negative breast cancer.
Pern F, Bogdanova N, Schürmann P, Lin M, Ay A, Länger F, Hillemanns P, Christiansen H, Park-Simon TW, Dörk T. Pern F, et al. PLoS One. 2012;7(10):e47993. doi: 10.1371/journal.pone.0047993. Epub 2012 Oct 24. PLoS One. 2012. PMID: 23110154 Free PMC article.
References
- Wooster R, Weber BL. N Engl J Med. 2003;348:2339–2347. - PubMed
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Mol Cell. 2006;22:719–729. - PubMed
- Patel KJ, Vu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, Colledge WH, Friedman LS, Ponder BA, Venkitaraman AR. Mol Cell. 1998;1:347–357. - PubMed
- Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, et al. Nat Genet. 2007;39:162–164. - PubMed
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. Nat Genet. 2007;39:159–161. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous