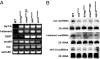Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster - PubMed (original) (raw)
Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster
Ai Khim Lim et al. Proc Natl Acad Sci U S A. 2007.
Erratum in
- Proc Natl Acad Sci U S A. 2007 Dec 11;104(50):20143
Abstract
The nuage is an electron-dense perinuclear structure that is known to be a hallmark of animal germ-line cells. Although the conservation of the nuage throughout evolution accentuates its essentiality, its role(s) and the exact mechanism(s) by which it functions in the germ line still remain unknown. Here, we report a nuage component, Krimper (KRIMP), in Drosophila melanogaster and show that it ensures the repression of the selfish genetic elements in the female germ line. The Krimp loss-of-function allele exhibited female sterility, defects in karyosome formation and oocyte polarity, and precocious osk translation. These phenotypes are commonly observed in the other nuage component mutants, vasa (vas) and maelstrom (mael), and the RNA-silencing component mutants, spindle-E (spn-E) and aubergine (aub), suggesting a shared underlying defect that uses RNA silencing. Moreover, we demonstrated that the localization of the nuage components depends on both SPN-E and AUB and that the selfish genetic elements were derepressed to different extents in the nuage component mutants, as well as in aub and armitage (armi) mutants. In the nuage component mutants, vas, krimp, and mael, the levels of roo, I-element, and HeT-A repeat-associated small interfering RNAs were greatly reduced. Hence, our data suggest that the nuage functions as a specialized center that protects the genome in the germ-line cells via gene regulation mediated by repeat-associated small interfering RNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
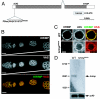
KRIMP is a nuage component. (A) Gene structure of krimp is shown. krimp contains two exons and is predicted to contain a tudor domain. A piggyBac insertion, f06583, 35 bp upstream of the krimp ORF results in female sterility. (B) KRIMP localizes to the perinuclear regions of the germ-line cells in the Drosophila ovary. Ovaries were immunostained with anti-KRIMP (shown in green) and anti-VAS (shown in red). KRIMP perinuclear foci colocalize with VAS foci. All ovarioles are orientated with the anterior to the left. (Scale bars: 20 μm.) (C) Closer view of a nurse cell nucleus confirms the colocalization. (Scale bars: 4 μm.) (D) _krimp_f06583 is a loss-of-function allele. Northern analysis indicates the expected transcript size of ≈2.5 kb in the WT ovaries. No detectable transcript is observed in krimp mutant ovaries.
Fig. 2.
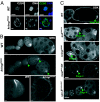
krimp mutants exhibit defective karyosome formation and oocyte polarity. (A) Immunostaining with a synaptonemal complex marker, C(3)G (shown in green), shows that the _krimp_f06583 oocyte nucleus fails to compact into a karyosome (shown in blue). (Scale bars: 5 μm.) (B) Ovary staining with anti-GRK. The level of GRK expression is down-regulated (indicated by green arrowheads), and its dorsal-anterior localization is disrupted in stage-8 egg chamber. (Scale bars: 20 μm.) (C) Ovary staining with anti-OSK. In WT, osk is translated at stages 7–9. Precocious translation of osk mRNA is observed in the nuage component mutants, including spn-E mutants, as indicated by green arrowheads. (Scale bars: 20 μm.)
Fig. 3.
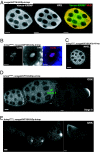
The UASp-venus(YFP)-krimp transgene fully rescues krimp mutant defects. Transgenic flies carrying the venus-krimp transgene under the UASp promoter were crossed with flies harboring the nosgal4VP16 transgene to drive the expression of Venus-tagged KRIMP in the ovary. (A) Immunostaining with anti-GFP (shown in green) and anti-VAS (shown in red) indicates that Venus-tagged KRIMP protein localizes to the perinuclear regions of the germ-line cells, which colocalize with VAS perinuclear foci. (Scale bars: 10 μm.) (B) By crossing flies harboring the venus-Krimp transgene into the _krimp_f06583 mutant background, the oocyte nucleus compacts into a karyosome (shown in blue), and C(3)G (shown in red) becomes chromosomal. (Scale bars: 5 μm.) (C) MAEL, whose localization depends on KRIMP, localizes to the perinuclear regions of the germ-line cells. (Scale bar: 10 μm.) (D) osk mRNA is translationally repressed normally. (Scale bars: 20 μm.) (E) GRK expression is comparable with the WT ovariole, and the protein localizes to the anterior-dorsal region of the stage-8 egg chamber. (Scale bars: 20 μm.)
Fig. 4.
Nuage foci are mislocalized in the germ-line-specific RNA-silencing component mutants. Ovaries from different mutant flies were immunostained for the nuage components. Homozygous mutant alleles or allelic combinations were used for all of the mutants, except for dcr-1, where clonal analysis was used. (A) Localization of the nuage components at the perinuclear regions of the germ-line cells reflects a hierarchical assembly. All of the nuage components, VAS, AUB, KRIMP, and MAEL, depend on SPN-E to localize normally to the perinuclear regions. AUB, KRIMP, and MAEL depend on VAS for proper localization; KRIMP and MAEL depend on VAS and AUB to localize to the nuage; MAEL depends on VAS, AUB, and KRIMP to localize normally. (B) Nuage localization is unaffected in the conventional dicing enzyme mutants, dcr-1 and dcr-2. (Scale bars: 10 μm.)
Fig. 5.
LINEs are derepressed, and the production of rasiRNAs is defective in the nuage component mutants. (A) The expression of the selfish genetic elements was examined by using semiquantitative RT-PCR. HeT-A and I-element were derepressed in the nuage component mutants, krimp and mael, and RNA-silencing component mutants, aub and armi. TART was derepressed in mael, aub, and armi mutants. mst40 was derepressed in krimp and mael mutants. roo, belonging to the euchromatic LTR family, was not derepressed. (B) PAGE northern analysis of roo, I-element, and HeT-A rasiRNAs. The production of roo, I-element, and HeT-A rasiRNAs was reduced in the nuage component mutants, as well as the RNA-silencing component mutants.
Similar articles
- Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas.
Patil VS, Kai T. Patil VS, et al. Curr Biol. 2010 Apr 27;20(8):724-30. doi: 10.1016/j.cub.2010.02.046. Epub 2010 Apr 1. Curr Biol. 2010. PMID: 20362446 - A novel organelle, the piNG-body, in the nuage of Drosophila male germ cells is associated with piRNA-mediated gene silencing.
Kibanov MV, Egorova KS, Ryazansky SS, Sokolova OA, Kotov AA, Olenkina OM, Stolyarenko AD, Gvozdev VA, Olenina LV. Kibanov MV, et al. Mol Biol Cell. 2011 Sep;22(18):3410-9. doi: 10.1091/mbc.E11-02-0168. Epub 2011 Jul 20. Mol Biol Cell. 2011. PMID: 21775629 Free PMC article. - [The interplay of transposon silencing genes in the Drosophila melanogaster germline].
Sokolova OA, Iakushev EIu, Stoliarenko AD, Mikhaleva EA, Gvozdev VA, Klenov MS. Sokolova OA, et al. Mol Biol (Mosk). 2011 Jul-Aug;45(4):633-41. Mol Biol (Mosk). 2011. PMID: 21954595 Russian. - piRNA pathway and the potential processing site, the nuage, in the Drosophila germline.
Pek JW, Patil VS, Kai T. Pek JW, et al. Dev Growth Differ. 2012 Jan;54(1):66-77. doi: 10.1111/j.1440-169x.2011.01316.x. Dev Growth Differ. 2012. PMID: 23741748 Review. - Factories without walls: The molecular architecture and functions of non-membrane organelles in small RNA-guided genome protection.
Kawaguchi S, Isshiki W, Kai T. Kawaguchi S, et al. Biochim Biophys Acta Gen Subj. 2025 Jun;1869(7):130811. doi: 10.1016/j.bbagen.2025.130811. Epub 2025 May 3. Biochim Biophys Acta Gen Subj. 2025. PMID: 40319768 Review.
Cited by
- UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery.
Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, Weng Z, Theurkauf WE. Zhang F, et al. Cell. 2012 Nov 9;151(4):871-884. doi: 10.1016/j.cell.2012.09.040. Cell. 2012. PMID: 23141543 Free PMC article. - Cytoplasmic compartmentalization of the fetal piRNA pathway in mice.
Aravin AA, van der Heijden GW, Castañeda J, Vagin VV, Hannon GJ, Bortvin A. Aravin AA, et al. PLoS Genet. 2009 Dec;5(12):e1000764. doi: 10.1371/journal.pgen.1000764. Epub 2009 Dec 11. PLoS Genet. 2009. PMID: 20011505 Free PMC article. - Roles of Piwi proteins in transcriptional regulation mediated by HP1s in cultured silkworm cells.
Tatsuke T, Zhu L, Li Z, Mitsunobu H, Yoshimura K, Mon H, Lee JM, Kusakabe T. Tatsuke T, et al. PLoS One. 2014 Mar 17;9(3):e92313. doi: 10.1371/journal.pone.0092313. eCollection 2014. PLoS One. 2014. PMID: 24637637 Free PMC article. - piRNA silencing contributes to interspecies hybrid sterility and reproductive isolation in Drosophila melanogaster.
Kotov AA, Adashev VE, Godneeva BK, Ninova M, Shatskikh AS, Bazylev SS, Aravin AA, Olenina LV. Kotov AA, et al. Nucleic Acids Res. 2019 May 7;47(8):4255-4271. doi: 10.1093/nar/gkz130. Nucleic Acids Res. 2019. PMID: 30788506 Free PMC article. - The evolution of RNAi as a defence against viruses and transposable elements.
Obbard DJ, Gordon KH, Buck AH, Jiggins FM. Obbard DJ, et al. Philos Trans R Soc Lond B Biol Sci. 2009 Jan 12;364(1513):99-115. doi: 10.1098/rstb.2008.0168. Philos Trans R Soc Lond B Biol Sci. 2009. PMID: 18926973 Free PMC article. Review.
References
- Eddy EM. Int Rev Cytol. 1975;43:229–281. - PubMed
- Liang L, Diehl-Jones W, Lasko P. Development (Cambridge, UK) 1994;120:1201–1211. - PubMed
- Harris AN, Macdonald PM. Development (Cambridge, UK) 2001;128:2823–2832. - PubMed
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Development (Cambridge, UK) 2003;130:859–871. - PubMed
- Kuznicki KA, Smith PA, Leung-Chiu WMA, Estevez AO, Scott HC, Bennett KL. Development (Cambridge, UK) 2000;127:2907–2916. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases