Cross-species difference in telomeric function of tankyrase 1 - PubMed (original) (raw)
Cross-species difference in telomeric function of tankyrase 1
Yukiko Muramatsu et al. Cancer Sci. 2007 Jun.
Abstract
Telomeres protect chromosome ends from being recognized as DNA double-strand breaks. Telomere shortening, which occurs due to incomplete replication of DNA termini, limits the proliferative capacity of human somatic cells and contributes as a barrier to carcinogenesis. In most human cancer cells, telomerase maintains telomere length whereas TRF1, a telomeric protein, represses telomere access to telomerase. Tankyrase 1 is a PARP that dissociates TRF1 from telomeres by poly(ADP-ribosyl)ating TRF1. Thus, by reducing TRF1 loading on chromosome ends, tankyrase 1 enhances telomere access to telomerase and causes telomere elongation. Recent studies of knockout mice suggest that tankyrases may not regulate telomere length in mice (Mus musculus). Consistent with this idea is that mouse TRF1 has no canonical tankyrase-binding motif. However, the presence of such a motif is not a prerequisite to bind tankyrase 1 in certain species. Here, we found that, in mice, tankyrase 1 does not bind or poly(ADP-ribosyl)ate TRF1. Accordingly, mouse TRF1 was resistant to tankyrase 1-mediated release from telomeres. These observations indicate that telomeric function of tankyrase 1 is not conserved in mice. We also found that the canonical tankyrase 1-binding motif in TRF1 is conserved in several mammals but not in rats. Since mice and rats have much higher telomerase activity in their somatic tissues and much longer telomeres than those in other mammals, these rodent species might have evolved to resign the tankyrase 1-mediated telomere maintenance system. Meanwhile, PARP inhibitors induced non-telomeric tankyrase 1 foci in the nuclei, suggesting another function of tankyrase 1 at non-telomeric loci.
Figures
Figure 1
Mouse and human tankyrase 1 show similar structural properties and expression patterns. (a) Top; identity between mouse and human tankyrase 1 amino acid sequences. HPS, homopolymeric runs of histidine, proline and serine; ANK, ankyrin domain, consisting of 24 ANK repeats; SAM, multimerization domain homologous to the sterile alpha motif; PARP, poly (ADP‐ribose) polymerase catalytic domain; ARC, ANK repeat cluster (see below). Bridges above two adjacent ANK repeats indicate the presence of a conserved histidine contributing to inter‐repeat stabilization. Bottom; tankyrase 1 ANK domain is divided into five well‐conserved ARCs. Two‐dimensional homology plot between 24 ANK repeats of ankyrin R and mouse full‐length tankyrase 1 (left) or full‐length‐tankyrase 1 and itself (right). Plots were made with DNASIS Version 3.5 (Hitachi Software); check size = 10; matching size = 5. Matching between equivalent amino acids (e.g. Ser and Thr) was not considered. (b) Northern blot analysis of mouse tankyrase 1. Multiple tissue northern blot (BD Clontech) was hybridized with 32P‐labeled tankyrase 1 HPS probe, which was not cross‐reacted with tankyrase 2 transcripts. Asterisk (*) indicates specific signal. Lower panel indicates the blot's calibration with a β‐actin probe. RNA size markers are indicated (left). (c) Subcellular fractionation of tankyrase 1 in mouse NIH3T3 cells. Each fraction was prepared as described in Materials and Methods and subjected to western blot analysis with primary antibodies indicated. Lane 1, cytoplasmic; lane 2, organelle/membrane; lane 3, nuclear; lane 4, cytoskeleton fractions. Asterisk (*) indicates specific signal. Blots with anti‐calpain I, GM130, histone H2AX, and vimentin antibodies demonstrate the purity of respective fractions.
Figure 2
Tankyrase 1 has no physical interaction with mouse telomeric repeat binding factor 1 (TRF1). (a) In vitro pull‐down assay of tankyrase 1 and TRF1. Human (left upper panel) and mouse (left lower panel) tankyrase 1 were prepared by in vitro translation and incubated with glutathione beads coupled with glutathione S‐transferase (GST)‐fused proteins as indicated. Coomassie Brilliant Blue (CBB)‐stained gel corresponding to the left upper panel is shown in the right panel. The bead‐bound tankyrase 1 was detected with western blot analysis. Molecular mass markers (K) are indicated at the left. (b) Partial amino acid sequences of human and mouse tankyrase‐binding protein of 182 kDa (TAB182). Each protein has a canonical tankyrase 1 binding motif, RPQPDG. Numbers indicate the positions of amino acid residues in the human paralog. (c) In vitro pull‐down assay of tankyrase 1 and TAB182. Human (H) and mouse (M) tankyrase 1 were prepared and incubated with glutathione beads coupled with GST‐fused proteins, as indicated. The bead‐bound tankyrase 1 was detected as in (a). (d) In vitro poly (ADP‐ribose) polymerase (PARP) assay. Tankyrase 1 was affinity‐purified from HTC75/FN‐tankyrase 1 (FLAG‐nuclear localization signal [NLS]‐tankyrase 1) cells( 13 ) and subjected to the PARP assay described in Materials and Methods. GST‐fused mouse and human TRF1 were used as acceptors for tankyrase 1's substrate, [32P]‐nicotinamide adenine dinucleotide (NAD). Reactions were carried out in the absence or the presence of 3 mM 3‐aminobenzamide (3AB), a PARP inhibitor, fractionated on sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE), and visualized by CBB staining (upper panel) or autoradiography (lower panel). Note that tankyrase 1 is also ADP‐ribosylated by itself.( 6 ) IP, immunoprecipitated; IgH, immunoglobulin heavy chain. (e) Co‐immunoprecipitation assay. HeLa I.2.11 cells were transiently transfected with Myc‐mTRF1 or Myc‐hTRF1. The cell lysates were prepared and immunoprecipitated with anti‐Myc antibody (9E10). The immunocomplex was subjected to western blot analysis with anti‐tankyrase antibody (H‐350).
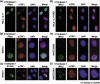
Figure 3
Tankyrase 1 dissociates telomeric repeat binding factor 1 (TRF1) from telomeres in humans but not in mice. (a) TRF1 release assay. Human HeLa I.2.11 cells were transfected with mouse MN‐tankyrase 1 (Myc‐NLS‐tankyrase 1). After a 20‐h incubation, cells were fixed with paraformaldehyde. MN‐tankyrase 1 and TRF1 were detected by indirect immunofluorescence stain with anti‐Myc (green) and anti‐human TRF1 5747 (red) antibodies, respectively. 4,6‐Diamino‐2‐phenylindole (DAPI) staining of DNA is shown in blue. (b, c) Mouse NIH3T3 cells were transfected with mouse MN‐tankyrase 1 (b) or human FN‐tankyrase 1 (c). The exogenous tankyrase 1 and the endogenous TRF1 were detected as in (a) with anti‐Myc/FLAG (green) and anti‐mouse TRF1 (red) antibodies, respectively. (d, e) HeLa I.2.11 (d) or NIH3T3 (e) cells were transfected with mouse MN‐tankyrase 1. After a 20‐h incubation in the presence of 3 mM 3AB, MN‐tankyrase 1 (green) and TRF1 (red) were detected. (f) NIH3T3 cells were transfected and incubated as in (e). Before fixation, Triton X‐100 extraction was carried out to eliminate DNA‐ and nuclear matrix‐unbound proteins. MN‐tankyrase 1 (green) and TRF1 (red) were detected.
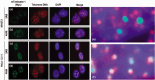
Figure 4
Poly (ADP‐ribose) polymerase (PARP) inhibitors reveal non‐telomeric tankyrase 1 foci in mouse and human cell nuclei. (a–d) NIH3T3 (a, b) or HeLa I.2.11 (c, d) cells were transfected with mouse MN‐tankyrase 1. After a 20‐h incubation in the presence of 3 mM 3‐aminobenzamide (3AB) (a, c) or 10 µM 4ANI (b, d), cells were fixed and MN‐tankyrase 1 (green) and telomeric DNA (red) were detected by immunofluorescence in situ hybridization (iFISH). (e, f) Magnified representation of (a) and (c), respectively.
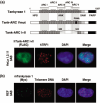
Figure 5
Characterization of non‐telomeric tankyrase 1 foci. (a) Formation of non‐telomeric tankyrase 1 foci depends on ankyrin repeat cluster (ARC) subdomains. Top; schematic view of a FN‐tankyrase 1 point mutant in which ARC V cannot bind TRF1 (FN‐tank‐ARC Vmut) and a deletion mutant that lacks functional ARCs III–V (FN‐tank‐ARC I + II).( 13 ) These constructs contain a FLAG tag and a nuclear localization signal (NLS) at the N‐termini (not shown). Numbers indicate the positions of amino acid residues. Bottom; HeLa I.2.11 cells were transfected with FN‐tank‐ARC I + II. After a 20‐h incubation in the presence of 3 mM 3‐aminobenzamide (3AB), the mutant FN‐tankyrase 1 (green) and TRF1 (red) were detected by indirect immunofluorescence staining. (b) REF (rat cells) were transfected with mouse MN‐tankyrase 1. After a 20‐h incubation in the presence of 3 mM 3AB, cells were fixed and MN‐tankyrase 1 (green) and telomeric DNA (red) were detected by immunofluorescence in situ hybridization (iFISH).
Similar articles
- Tankyrase promotes telomere elongation in human cells.
Smith S, de Lange T. Smith S, et al. Curr Biol. 2000 Oct 19;10(20):1299-302. doi: 10.1016/s0960-9822(00)00752-1. Curr Biol. 2000. PMID: 11069113 - Telomere elongation by a mutant tankyrase 1 without TRF1 poly(ADP-ribosyl)ation.
Muramatsu Y, Tahara H, Ono T, Tsuruo T, Seimiya H. Muramatsu Y, et al. Exp Cell Res. 2008 Mar 10;314(5):1115-24. doi: 10.1016/j.yexcr.2007.12.005. Epub 2007 Dec 14. Exp Cell Res. 2008. PMID: 18221737 - Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres.
Cook BD, Dynek JN, Chang W, Shostak G, Smith S. Cook BD, et al. Mol Cell Biol. 2002 Jan;22(1):332-42. doi: 10.1128/MCB.22.1.332-342.2002. Mol Cell Biol. 2002. PMID: 11739745 Free PMC article. - [The role of telomere-binding proteins in carcinogenesis].
Aragona M, Pontoriero A, Panetta S, La Torre I, La Torre F. Aragona M, et al. Minerva Med. 2000 Nov-Dec;91(11-12):299-304. Minerva Med. 2000. PMID: 11253711 Review. Italian. - Post-translational modifications of TRF1 and TRF2 and their roles in telomere maintenance.
Walker JR, Zhu XD. Walker JR, et al. Mech Ageing Dev. 2012 Jun;133(6):421-34. doi: 10.1016/j.mad.2012.05.002. Epub 2012 May 23. Mech Ageing Dev. 2012. PMID: 22634377 Review.
Cited by
- Telomestatin impairs glioma stem cell survival and growth through the disruption of telomeric G-quadruplex and inhibition of the proto-oncogene, c-Myb.
Miyazaki T, Pan Y, Joshi K, Purohit D, Hu B, Demir H, Mazumder S, Okabe S, Yamori T, Viapiano M, Shin-ya K, Seimiya H, Nakano I. Miyazaki T, et al. Clin Cancer Res. 2012 Mar 1;18(5):1268-80. doi: 10.1158/1078-0432.CCR-11-1795. Epub 2012 Jan 9. Clin Cancer Res. 2012. PMID: 22230766 Free PMC article. - An Evolutionary Perspective on the Origin, Conservation and Binding Partner Acquisition of Tankyrases.
Sowa ST, Bosetti C, Galera-Prat A, Johnson MS, Lehtiö L. Sowa ST, et al. Biomolecules. 2022 Nov 15;12(11):1688. doi: 10.3390/biom12111688. Biomolecules. 2022. PMID: 36421702 Free PMC article. - Human tankyrases are aberrantly expressed in colon tumors and contain multiple epitopes that induce humoral and cellular immune responses in cancer patients.
Shebzukhov YV, Lavrik IN, Karbach J, Khlgatian SV, Koroleva EP, Belousov PV, Kashkin KN, Knuth A, Jager E, Chi NW, Kuprash DV, Nedospasov SA. Shebzukhov YV, et al. Cancer Immunol Immunother. 2008 Jun;57(6):871-81. doi: 10.1007/s00262-007-0423-z. Epub 2007 Nov 20. Cancer Immunol Immunother. 2008. PMID: 18026951 Free PMC article. - Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice.
Martínez P, Thanasoula M, Muñoz P, Liao C, Tejera A, McNees C, Flores JM, Fernández-Capetillo O, Tarsounas M, Blasco MA. Martínez P, et al. Genes Dev. 2009 Sep 1;23(17):2060-75. doi: 10.1101/gad.543509. Epub 2009 Aug 13. Genes Dev. 2009. PMID: 19679647 Free PMC article. - Knockdown of tankyrase 1 inhibits the progression of gastric adenocarcinoma via regulating human telomerase reverse transcriptase and telomeric repeat binding factor 1.
Liu W, Zhu JJ, Liu YY, Tang NN, Wang Y, Jiang B, Wang LL, Wang YX, Xie MP, Wang XY. Liu W, et al. J Gastrointest Oncol. 2022 Apr;13(2):559-568. doi: 10.21037/jgo-22-82. J Gastrointest Oncol. 2022. PMID: 35557584 Free PMC article.
References
- De Lange T. Mammalian telomeres. In: De Lange T, Lundblad V, Blackburn EH, eds. Telomeres, 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 2006; 387–431.
- D’Adda di Fagagna F, Reaper PM, Clay‐Farrace L et al. A DNA damage checkpoint response in telomere‐initiated senescence. Nature 2003; 426: 194–8. - PubMed
- Kim NW, Piatyszek MA, Prowse KR et al. Specific association of human telomerase activity with immortal cells and cancer. Science 1994; 266: 2011–15. - PubMed
- Bodnar AG, Ouellette M, Frolkis M et al. Extension of life‐span by introduction of telomerase into normal human cells. Science 1998; 279: 349–52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials