The structural determinants of checkpoint activation - PubMed (original) (raw)
The structural determinants of checkpoint activation
Christina A MacDougall et al. Genes Dev. 2007.
Abstract
Here, we demonstrate that primed, single-stranded DNA (ssDNA) is sufficient for activation of the ATR-dependent checkpoint pathway in Xenopus egg extracts. Using this structure, we define the contribution of the 5'- and 3'-primer ends to Chk1 activation when replication is blocked and ongoing. In addition, we show that although ssDNA is not sufficient for checkpoint activation, the amount of ssDNA adjacent to the primer influences the level of Chk1 phosphorylation. These observations define the minimal DNA requirements for checkpoint activation and suggest that primed ssDNA represents a common checkpoint activating-structure formed following many types of damage.
Figures
Figure 1.
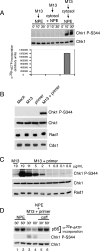
Primed M13 ssDNA induces Chk1 phosphorylation and checkpoint activation. (A) M13 ssDNA (30 μg/mL) was added to NPE, a 1:1 mixture of cytosol and NPE, or cytosol (30 min) and then NPE. Parallel samples were removed at the indicated times post-DNA addition for NPE and cytosol + NPE. For the sequential addition, times were post-NPE addition. Samples were either immunoblotted for phospho-Chk1 (S344) and Chk1 (top panels), or replication was monitored by measuring α32P-dCTP incorporation following gel electrophoresis (bottom). (B) Buffer, M13 ssDNA, the ssDNA primer (80-mer), or M13 preannealed to the primer (M13 + primer) was added to NPE. Samples were taken after 20 min and analyzed by immunoblotting after SDS-PAGE with antibodies for phospho-Chk1 (S344), Chk1, Rad1, and Cds1. (C) ssM13 DNA or M13 + primer was added to NPE at the indicated concentrations. Samples were analyzed as in B. (D) Double-stranded pBS (30 μg/mL) was incubated in cytosol for 30 min and added to an equal volume of NPE (lanes 1,2), NPE containing M13 + primer (20 min; 12.5 μg/mL; lanes 3,4), or NPE containing M13 + primer + caffeine (20 min; 12.5 μg/mL; caff, 4 mM; lanes 5,6). Parallel samples were removed 30 and 60 min post-addition of plasmid to NPE and analyzed for replication as in A (top panel) or immunoblotted for phospho-Chk1 (S344) and Chk1 (bottom panels). Final concentrations are shown.
Figure 2.
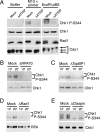
Effect of checkpoint protein depletion on phosphorylation events induced by primed M13 ssDNA. (A) Buffer, M13 + primer, or EcoRI-digested pBS (0.2 μg/mL) was added to mock-depleted or ATRIP-depleted NPE, and samples were analyzed as in Figure 1B. (B_–_E) NPE was mock-depleted or depleted with antibodies to RPA70 (B), TopBP1 (C), Rad1 (D), or Claspin (E). Mock-depleted NPE or depleted NPE was then incubated with M13 + primer, and samples were analyzed as in Figure 1B. Chk1 (B,C,E) and RPA (D) levels were monitored as loading controls. The lower band in E represents cross-reacting residual IgG in the extract from depletion.
Figure 3.
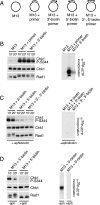
Effect of modifying primer ends on Chk1 and Rad1 phosphorylation induced by primed M13 ssDNA. (A) Names and figures of structures used in B_–_D. Dot represents the biotinylated end of the primer. (B) M13, M13 + unmodified 80-mer, or M13 + 80-mer blocked at the 3′-end with biotin was incubated with streptavidin and added to NPE. Samples were taken and analyzed for phospho-Chk1 (S344), Chk1, or Rad1 by immunoblotting, or for replication as described in Figure 1A. (C) M13 ssDNA annealed to an 80-mer blocked at the 3′-end, 5′-end, or 3′- and 5′-ends with biotin were incubated with streptavidin, added to NPE in the presence of aphidicolin, and analyzed as described in B. (D) M13 ssDNA annealed to an 80-mer blocked at the 5′-end with biotin was incubated with streptavidin, added to NPE in the absence or presence of aphidicolin, and analyzed as described in B.
Figure 4.
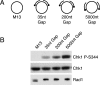
Effect of ssDNA gap size on Chk1 phosphorylation. (A) Names and figures of structures used in B. Dot represents the biotinylated end of the primer. (B) The structures shown were incubated with streptavidin and then added to NPE in the presence of aphidicolin. Samples were analyzed as in Figure 3B.
Comment in
- Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response.
Zou L. Zou L. Genes Dev. 2007 Apr 15;21(8):879-85. doi: 10.1101/gad.1550307. Genes Dev. 2007. PMID: 17437994 No abstract available.
Similar articles
- APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress.
Willis J, Patel Y, Lentz BL, Yan S. Willis J, et al. Proc Natl Acad Sci U S A. 2013 Jun 25;110(26):10592-7. doi: 10.1073/pnas.1301445110. Epub 2013 Jun 10. Proc Natl Acad Sci U S A. 2013. PMID: 23754435 Free PMC article. - Phosphorylation of replication protein A by S-phase checkpoint kinases.
Liu JS, Kuo SR, Melendy T. Liu JS, et al. DNA Repair (Amst). 2006 Mar 7;5(3):369-80. doi: 10.1016/j.dnarep.2005.11.007. Epub 2006 Jan 18. DNA Repair (Amst). 2006. PMID: 16412704 - Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint.
Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Byun TS, et al. Genes Dev. 2005 May 1;19(9):1040-52. doi: 10.1101/gad.1301205. Epub 2005 Apr 15. Genes Dev. 2005. PMID: 15833913 Free PMC article. - Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase.
Petermann E, Caldecott KW. Petermann E, et al. Cell Cycle. 2006 Oct;5(19):2203-9. doi: 10.4161/cc.5.19.3256. Epub 2006 Oct 1. Cell Cycle. 2006. PMID: 16969104 Review. - Chk1 in the DNA damage response: conserved roles from yeasts to mammals.
Chen Y, Sanchez Y. Chen Y, et al. DNA Repair (Amst). 2004 Aug-Sep;3(8-9):1025-32. doi: 10.1016/j.dnarep.2004.03.003. DNA Repair (Amst). 2004. PMID: 15279789 Review.
Cited by
- DEAD Box 1 Facilitates Removal of RNA and Homologous Recombination at DNA Double-Strand Breaks.
Li L, Germain DR, Poon HY, Hildebrandt MR, Monckton EA, McDonald D, Hendzel MJ, Godbout R. Li L, et al. Mol Cell Biol. 2016 Oct 28;36(22):2794-2810. doi: 10.1128/MCB.00415-16. Print 2016 Nov 15. Mol Cell Biol. 2016. PMID: 27550810 Free PMC article. - Role of replication protein A as sensor in activation of the S-phase checkpoint in Xenopus egg extracts.
Recolin B, Van der Laan S, Maiorano D. Recolin B, et al. Nucleic Acids Res. 2012 Apr;40(8):3431-42. doi: 10.1093/nar/gkr1241. Epub 2011 Dec 19. Nucleic Acids Res. 2012. PMID: 22187152 Free PMC article. - ATR preferentially interacts with common fragile site FRA3B and the binding requires its kinase activity in response to aphidicolin treatment.
Wan C, Kulkarni A, Wang YH. Wan C, et al. Mutat Res. 2010 Apr 1;686(1-2):39-46. doi: 10.1016/j.mrfmmm.2009.12.012. Epub 2010 Jan 7. Mutat Res. 2010. PMID: 20060399 Free PMC article. - Generation and Analysis of dsDNA Breaks for Checkpoint and Repair Studies in Fission Yeast.
Ramalingam R, O'Connell MJ. Ramalingam R, et al. Methods Mol Biol. 2021;2267:191-205. doi: 10.1007/978-1-0716-1217-0_13. Methods Mol Biol. 2021. PMID: 33786793 - Separation of intra-S checkpoint protein contributions to DNA replication fork protection and genomic stability in normal human fibroblasts.
Smith-Roe SL, Patel SS, Zhou Y, Simpson DA, Rao S, Ibrahim JG, Cordeiro-Stone M, Kaufmann WK. Smith-Roe SL, et al. Cell Cycle. 2013 Jan 15;12(2):332-45. doi: 10.4161/cc.23177. Epub 2012 Jan 15. Cell Cycle. 2013. PMID: 23255133 Free PMC article.
References
- Ahn J., Urist M., Prives C., Urist M., Prives C., Prives C. The Chk2 protein kinase. DNA Repair (Amst.) 2004;3:1039–1047. - PubMed
- Byun T.S., Pacek M., Yee M.C., Walter J.C., Cimprich K.A., Pacek M., Yee M.C., Walter J.C., Cimprich K.A., Yee M.C., Walter J.C., Cimprich K.A., Walter J.C., Cimprich K.A., Cimprich K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes & Dev. 2005;19:1040–1052. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous