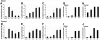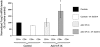Induction and role of regulatory CD4+CD25+ T cells in tolerance to the transgene product following hepatic in vivo gene transfer - PubMed (original) (raw)
Induction and role of regulatory CD4+CD25+ T cells in tolerance to the transgene product following hepatic in vivo gene transfer
Ou Cao et al. Blood. 2007.
Abstract
Gene replacement therapy is complicated by the risk of an immune response against the therapeutic transgene product, which in part is determined by the route of vector administration. Our previous studies demonstrated induction of immune tolerance to coagulation factor IX (FIX) by hepatic adeno-associated viral (AAV) gene transfer. Using a regulatory T-cell (T(reg))-deficient model (Rag-2(-/-) mice transgenic for ovalbumin-specific T-cell receptor DO11.10), we provide first definitive evidence for induction of transgene product-specific CD4(+)CD25(+) T(regs) by in vivo gene transfer. Hepatic gene transfer-induced T(regs) express FoxP3, GITR, and CTLA4, and suppress CD4(+)CD25(-) T cells. T(regs) are detected as early as 2 weeks after gene transfer, and increase in frequency in thymus and secondary lymphoid organs during the following 2 months. Similarly, adoptive lymphocyte transfers from mice tolerized to human FIX by hepatic AAV gene transfer indicate induction of CD4(+)CD25(+)GITR(+) that suppresses antibody formation to FIX. Moreover, in vivo depletion of CD4(+)CD25(+) T(regs) leads to antibody formation to the FIX transgene product after hepatic gene transfer, which strongly suggests that these regulatory cells are required for tolerance induction. Our study reveals a crucial role of CD4(+)CD25(+) T(regs) in preventing immune responses to the transgene product in gene transfer.
Figures
Figure 1
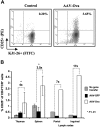
Percent CD25+ T cells of CD4+ DO11.10 TCR+ cells in DO11.10-tg RAG-2−/− BALB/c mice as determined by flow cytometry. (A) Scatter graphs show staining of CD25 and DO11.10 TCR for gated CD4+ cells. This example represents inguinal lymph node cells from control mice and animals that received hepatic gene transfer with 1 × 1012 vg AAV-EF1α-ova vector. Mice were killed 8 weeks after gene transfer. Antibody stainings were FITC-conjugated KJ1-26 (for DO11.10 TCR) and PE-conjugated CD25. (B) Summary of results for different lymphoid organs including fold difference between groups. Mice had received no gene transfer or AAV-EF1α-GFP or AAV-EF1α-ova. Results from spleens and thymus were average (± SD), while lymph nodes cells were pooled prior to flow cytometry (n = 5/cohort). *P < .01 compared with control groups.
Figure 2
Characterization of regulatory CD4+CD25+ cells isolated from AAV-EF1α-ova–transduced DO11.10-tg RAG-2−/− mice. Expression of FoxP3 in a subpopulation of CD4+ splenocytes derived from DO11.10-tg BALB/c (RAG+) or DO11.10-tg RAG-2−/− (RAG−) mice. (A) Splenocytes were sorted into CD4+ cells and further separated into CD25+ or CD25− cells. cDNA from each population was subjected to PCR using FoxP3- or HPRT (hypoxanthine–guanine phosphoribosyl–transferase)–specific primers. (B) Quantification of relative FoxP3 mRNA levels in indicated CD4+ T-cell subsets. cDNA samples were subjected to real-time quantitative PCR analyses using primers and an internal fluorescent probe specific for FoxP3 or HPRT. The relative quantity of FoxP3 in each sample was normalized to the relative quantity of HPRT. Shown are average results for 3 independent experiments (± SD). (C) Flow cytometry for FoxP3 expression. Shown are examples for percent CD25+FoxP3+ of CD4+ splenocytes in individual mice (8 weeks after gene transfer with AAV-ova or naive control). (D) Cell dose–dependent suppression of IL-2 expression in DO11.10-tg CD4+CD25− cells by CD4+CD25+ isolated from AAV-EF1α-ova–transduced DO11.10-tg RAG-2−/− mice upon in vitro coculture (in the presence of APCs) and stimulation with ova. Data represent average (± SD) from n = 4 cultures based on cells pooled from several mice.
Figure 3
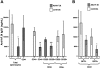
Analyses of cell populations from lymphoid organs of DO11.10-tg RAG-2−/− mice by flow cytometry 60 days after AAV-EF1α-ova gene transfer. (A) Quantitation of CD25+ of TCR+CD4+, CD25+GITR+ and CD25+CTLA4+ of CD4+ cells from spleen in naive or vector-treated mice. Percentages of dual-positive cells (top-right quadrant) are indicated. Note that AAV-EF1α-ova–transduced mice showed substantial increase in CD25+, CD25+GITR+, and CD25+CTLA4+ of total CD4+ cells compared with control animals. Antibody stain was FITC-conjugated KJ1-26 or CD4, PE-conjugated GITR or CTLA4, and APC-conjugated CD25. Representative examples of FACS density plots are shown for individual samples of splenocytes from DO11.10-tg RAG-2−/− mice. Also graphed are percent GITR+ (B) and CTLA4+ (C) of CD4+CD25+ cells derived from spleen, thymus, portal nodes, and inguinal nodes 60 days after AAV-EF1α-ova gene delivery (B,C).
Figure 4
Analyses of lymphocyte populations in DO11.10-tg RAG-2−/− mice as a function of time after administration of AAV-EF1α-ova. Examined tissues were spleens and thymus. Shown is the summary of percent CD25+ (A,D), CD25+GITR+ (B,E), and CD25+CTLA4+ (C,F) of total CD4+ cells derived from spleen or thymus. Note that more than 99% of CD4+ cells in these mice express ova-specific TCR DO11.10 and are therefore KJ1-26+. Antibody staining was carried out using FITC-conjugated KJ1-26 or CD4, PE-conjugated GITR or CTLA4, APC-conjugated CD25. Results are average ± SD for n = 3 mice. Also shown is a summary of percent GITR+ (G,H), and CTLA4+ (I,J) of CD4+CD25+ cells from spleen (G,I) or thymus (H,J) as a function of time after vector administration.
Figure 5
Plasma levels of IgG1 anti-hFIX 3 weeks after immunologic challenge by subcutaneous administration of 5 μg hFIX formulated in CFA in C57BL/6 mice that had received adoptive transfer of splenocytes from naive (white bars, controls) or vector-treated (hatched bars, AAV-FIX) C57BL/6 mice. Adoptive transfer was by tail vein injection 24 hours before challenge. Vector-treated mice had received hepatic gene transfer with 1011 vg/animal AAV-ApoE/hAAT-hFIX vector 6 weeks before the experiment. (A) Total splenocytes (5 × 107, bars 1-2), CD4+ T-cell–depleted splenocytes (5 × 107, bar 3), CD25+-depleted CD4+ T cells (9 × 106, bars 6, 8), MACS-purified CD4+ T cells (107, bars 4-5), or CD4+CD25+ cells (1 × 106, bars 7, 9) were transferred. Each bar is average antibody titer for 6 animals (± SD). *P < .05 compared with control splenocytes. **P < .01 compared with control CD4+ cells. ***P < .01 compared with CD4+CD25− cells or to control CD4+ cells. (B) CD4+GITR+ (bars 12-13) or CD4+GITR− splenocytes (bars 10-11) were transferred. Each bar is average antibody titer for 4 animals (± SD). *P < .05 compared with CD4+GITR− cells or to control CD4+GITR+ cells.
Figure 6
Quantification of relative FoxP3 mRNA levels in CD4+ or CD4− T cells derived from naive control or AAV-ApoE/hAAT-hFIX gene-transferred C57BL/6 mice 60 days before the experiment. Some of control or gene-transduced animals were challenged by subcutaneous administration of 5 μg hFIX formulated in CFA 5 days prior to PCR analysis. cDNA samples were subjected to real-time quantitative PCR analyses using primers and an internal fluorescent probe specific for FoxP3 or HPRT. The relative quantity of FoxP3 in each sample was normalized to the relative quantity of HPRT. FoxP3 transcript levels in naive CD4+ cells are reported as 100%. Results were shown as average ± SD (n = 3 per group). *P < .05 compared with other experimental groups.
Figure 7
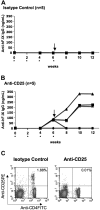
Depletion of CD4+CD25+ cells in C57BL/6 mice after gene transfer with AAV-ApoE/hAAT-hFIX vector. Shown are plasma levels of IgG anti-hFIX as a function of time after vector administration. Animals were injected intraperitoneally with either rat IgG1 (A, isotype control) or rat antimouse CD25 monoclonal antibody PC61 (B) every 2 weeks (up to week 6) starting on the day of liver-directed gene transfer (asterisk). Mice were challenged with 5 μg hFIX formulated in CFA at week 6. (C) Analyses of peripheral blood lymphocytes by flow cytometry showed depletion of CD4+CD25+ cells after repetitive anti-CD25 injection in C57BL/6 mice. Antibody staining was carried out using FITC-conjugated anti-CD4 and anti–PE-conjugated CD25.
Similar articles
- Induction of antigen-specific CD4+ T-cell anergy and deletion by in vivo viral gene transfer.
Dobrzynski E, Mingozzi F, Liu YL, Bendo E, Cao O, Wang L, Herzog RW. Dobrzynski E, et al. Blood. 2004 Aug 15;104(4):969-77. doi: 10.1182/blood-2004-03-0847. Epub 2004 Apr 22. Blood. 2004. PMID: 15105293 - Role of antigen-specific regulatory CD4+CD25+ T cells in tolerance induction after neonatal IP administration of AAV-hF.IX.
Shi Y, Falahati R, Zhang J, Flebbe-Rehwaldt L, Gaensler KM. Shi Y, et al. Gene Ther. 2013 Oct;20(10):987-96. doi: 10.1038/gt.2013.22. Epub 2013 Jun 13. Gene Ther. 2013. PMID: 23759700 Free PMC article. - GITR engagement preferentially enhances proliferation of functionally competent CD4+CD25+FoxP3+ regulatory T cells.
Liao G, Nayak S, Regueiro JR, Berger SB, Detre C, Romero X, de Waal Malefyt R, Chatila TA, Herzog RW, Terhorst C. Liao G, et al. Int Immunol. 2010 Apr;22(4):259-70. doi: 10.1093/intimm/dxq001. Epub 2010 Feb 5. Int Immunol. 2010. PMID: 20139172 Free PMC article. - Tolerance induction by viral in vivo gene transfer.
Dobrzynski E, Herzog RW. Dobrzynski E, et al. Clin Med Res. 2005 Nov;3(4):234-40. doi: 10.3121/cmr.3.4.234. Clin Med Res. 2005. PMID: 16303889 Free PMC article. Review. - Emerging role of regulatory T cells in gene transfer.
Cao O, Furlan-Freguia C, Arruda VR, Herzog RW. Cao O, et al. Curr Gene Ther. 2007 Oct;7(5):381-90. doi: 10.2174/156652307782151506. Curr Gene Ther. 2007. PMID: 17979684 Review.
Cited by
- Exploring the Role of GITR/GITRL Signaling: From Liver Disease to Hepatocellular Carcinoma.
Papadakos SP, Chatzikalil E, Vakadaris G, Reppas L, Arvanitakis K, Koufakis T, Siakavellas SI, Manolakopoulos S, Germanidis G, Theocharis S. Papadakos SP, et al. Cancers (Basel). 2024 Jul 22;16(14):2609. doi: 10.3390/cancers16142609. Cancers (Basel). 2024. PMID: 39061246 Free PMC article. Review. - Role of FoxP3+ Regulatory T Cells in Modulating Immune Responses to Adeno-Associated Virus Gene Therapy.
Muñoz-Melero M, Biswas M. Muñoz-Melero M, et al. Hum Gene Ther. 2024 Jul;35(13-14):439-450. doi: 10.1089/hum.2023.227. Epub 2024 Apr 11. Hum Gene Ther. 2024. PMID: 38450566 Review. - The novel mechanism facilitating chronic hepatitis B infection: immunometabolism and epigenetic modification reprogramming.
Wang Z, Liu N, Yang Y, Tu Z. Wang Z, et al. Front Immunol. 2024 Jan 15;15:1349867. doi: 10.3389/fimmu.2024.1349867. eCollection 2024. Front Immunol. 2024. PMID: 38288308 Free PMC article. Review. - AAV Immunotoxicity: Implications in Anti-HBV Gene Therapy.
Jacobs R, Dogbey MD, Mnyandu N, Neves K, Barth S, Arbuthnot P, Maepa MB. Jacobs R, et al. Microorganisms. 2023 Dec 14;11(12):2985. doi: 10.3390/microorganisms11122985. Microorganisms. 2023. PMID: 38138129 Free PMC article. Review. - TLR9-independent CD8+ T cell responses in hepatic AAV gene transfer through IL-1R1-MyD88 signaling.
Kumar SRP, Biswas M, Cao D, Arisa S, Muñoz-Melero M, Lam AK, Piñeros AR, Kapur R, Kaisho T, Kaufman RJ, Xiao W, Shayakhmetov DM, Terhorst C, de Jong YP, Herzog RW. Kumar SRP, et al. Mol Ther. 2024 Feb 7;32(2):325-339. doi: 10.1016/j.ymthe.2023.11.029. Epub 2023 Dec 5. Mol Ther. 2024. PMID: 38053332
References
- Cao O, Herzog RW, Hagstrom JN, Wang L. Gene therapy for treatment of inherited haematological disorders. Expert Opin Biol Ther. 2006;6:509–522. - PubMed
- Warrington KH, Jr, Herzog RW. Treatment of human disease by adeno-associated viral gene transfer. Hum Genet. 2006;119:571–603. - PubMed
- Herzog RW, Dobrzynski E. Immune implications of gene therapy for hemophilia. Semin Thromb Hemost. 2004;30:215–226. - PubMed
- Sabatino DE, High KA. Immune responses in gene transfer for genetic disorders. New York, NY: Kluwer Academic Press; p. 2002.
- Wang L, Cao O, Swalm B, Dobrzynski E, Mingozzi F, Herzog RW. Major role of local immune responses in antibody formation to factor IX in AAV gene transfer. Gene Ther. 2005;12:1453–1464. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous