Polycomb protein Cbx4 promotes SUMO modification of de novo DNA methyltransferase Dnmt3a - PubMed (original) (raw)
Polycomb protein Cbx4 promotes SUMO modification of de novo DNA methyltransferase Dnmt3a
Bing Li et al. Biochem J. 2007.
Abstract
The 'de novo methyltransferase' Dnmt3a (DNA methyltransferase 3a) has been shown to mediate transcriptional repression. Post-translational modification of Dnmt3a by SUMOylation affects its ability to transcriptionally repress. However, very little is known about how the SUMOylation process is regulated. In the present study, we identified a PcG (Polycomb group) protein, Cbx4 (chromobox 4), as a specific interaction partner of Dnmt3a. Co-expression of Cbx4 and SUMO-1 (small ubiquitin-related modifier-1) along with Dnmt3a in transfected cells results in enhanced modification of Dnmt3a with SUMO-1. Purified Cbx4 also promotes SUMOylation of Dnmt3a in vitro. The modification occurs in the N-terminal regulatory region, including the PWWP (Pro-Trp-Trp-Pro) domain. Our results suggest that Cbx4 functions as a SUMO E3 ligase for Dnmt3a and it might be involved in the functional regulation of DNA methyltransferases by promoting their SUMO modification.
Figures
Figure 1. Cbx4 interacts with Dnmt3a in vitro and in vivo
(A) Direct interaction of Cbx4 with Dnmt3a: Western-blotting analysis of the His-tagged Cbx4 in fractions obtained from GST pull-down assays using GST (lanes 2 and 5) or GST–Dnmt3a (lanes 3 and 6). About 10% of the input Cbx4 was loaded (lanes 1 and 4). To eliminate the possibility of false-positive interactions caused by contaminating nucleic acid, micrococcal nuclease (MNase) or ethidium bromide (EtBr) was used to treat the protein preparations during pull-down assays. (B) Co-immunoprecipitation (IP) of Dnmt3a with Cbx4 expressed from transfected cells. HEK-293T cells were transiently transfected with pcDNA3-HA-Dnmt3a and pFlag-CMV-Cbx4 alone or in combination as indicated at the top. Whole-cell extracts were immunoprecipitated with anti-Flag antibody. The precipitated proteins were analysed by Western blotting using anti-HA or anti-Flag antibody as indicated. (C) Co-immunoprecipitation of Cbx4 with Dnmt3a from transfected cells. Samples immunoprecipitated with anti-HA antibody were analysed by Western blotting using anti-Cbx4 or anti-HA antibody as indicated.
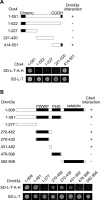
Figure 2. Mapping of interaction domains in Cbx4 and Dnmt3a
(A) Schematic representation of Cbx4 fragments with their Dnmt3a-interacting abilities (upper panel, shown on the right) based on the data from yeast two-hybrid assays (lower panel). The full-length Dnmt3a was fused to GAL4BD. The fusion construct was co-transformed into yeast strain AH109 with the indicated portions of Cbx4, which were fused to the GAL4AD. Growth of colonies on the SD minimal medium (SD-L-T-A-H) lacking leucine, tryptophan, adenine and histidine reflects positive interaction of Cbx4 fragments with Dnmt3a. Regions of conservation (the chromodomain and the COOH box) are depicted with black shading in the bars. (B) Schematic representation of Dnmt3a fragments with their Cbx4-interacting abilities (upper panel, shown on the right) based on the data from yeast two-hybrid assays (lower panel). Full-length Cbx4 was fused to GAL4AD and was co-transformed into yeast strain AH109 with the indicated portions of Dnmt3a, which are fused to the GAL4BD. Growth of colonies on the SD minimal medium (SD-L-T-A-H) reflects positive interactions.
Figure 3. Dnmt3a interacts with Cbx4 specifically
(A) Amino acid alignment of the COOH boxes and their flanking regions of the Cbx family members. Identical residues are indicated by black shading and highly conserved residues by grey shading. (B) Schematic representation of Cbx proteins with their Dnmt3a-interacting abilities (upper panel, shown on the right) based on the data from yeast two-hybrid assays (middle panel). Cbx proteins were fused to GAL4AD and co-transformed into yeast strain AH109 with GAL4BD–Dnmt3a. The interactions were accessed by growth of colonies on the SD minimal medium (SD-L-T-A-H). The GAL4AD-Cbx and GAL4BD-Dnmt3a fusion proteins also contain an HA or a myc epitope tag respectively, which were used to confirm the expression of Cbx proteins and Dnmt3a in transformed yeast by Western blotting (bottom panel).
Figure 4. Dnmt3a is SUMOylated in HEK-293T cells
(A) Western-blotting analysis of the SUMOylation of Dnmt3a. HEK-293T cells were transiently transfected with expression plasmids as indicated on the top. Cell lysates were analysed by Western blotting with anti-HA antibody (left panel) or with anti-Dnmt3a antibody (right panel). The positions of Dnmt3a and its SUMOylated forms are indicated. (B) Immunoprecipitation analysis of the SUMOylation of Dnmt3a. HEK-293T cells were transiently transfected with expression plasmids as indicated on the top. The cell extracts were immunoprecipitated with anti-HA antibody. The precipitated proteins were analysed by Western blotting with anti-Dnmt3a antibody (lanes 1 and 2), anti-GFP antibody (lanes 3 and 4) or anti-HA antibody (lanes 5 and 6). The positions of Dnmt3a and its SUMOylated forms are indicated.
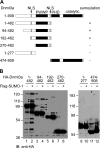
Figure 5. Mapping of the Dnmt3a region necessary for SUMOylation
(A) Schematic overview of the Dnmt3a deletion mutants used. The NLS sequences are indicated by arrows. The ability to be SUMOylated was assessed on the basis of the data in (B) and is indicated on the right. (B) Western-blotting analysis of the SUMOylation of Dnmt3a deletion mutants. HEK-293T cells were transiently transfected with expression plasmids encoding different regions of Dnmt3a, as indicated on the top. Cell lysates were analysed by Western blotting with anti-HA antibody. The positions of SUMOylated forms of Dnmt3a fragments are marked by arrowheads. Arrows depict the additional bands which might represent an unknown form of post-translational modification of Dnmt3a.
Figure 6. Cbx4 enhances SUMOylation of Dnmt3a in vivo and in vitro
(A) Cbx4 promotes SUMO-1 conjugation to CtBP in transfected cells. HEK-293T cells were transiently transfected with expression plasmids encoding Flag–SUMO-1, Flag–Cbx4 and myc-CtBP as indicated. The cell lysates were analysed by Western blotting using anti-myc (upper panel) or anti-Cbx4 (lower panel) antibody. (B) Cbx4 promotes SUMO-1 conjugation to Dnmt3a in transfected cells. HEK-293T cells were transiently transfected with expression plasmids encoding GFP–SUMO-1, Flag–Cbx4 and HA–Dnmt3a as indicated. The cell lysates were analysed by Western blotting using anti-HA (upper panel) or anti-Cbx4 antibody (lower panel). (C) SUMOylation of Dnmt3a in vitro. Purified GST-tagged Dnmt3a (amino acids 1–482, GST–Dnmt3aN) was incubated in the absence or presence of 250 ng of recombinant E1 (Aos1/Uba2 heterodimer), 100 ng of recombinant E2 (Ubc9) and 3 μg of recombinant SUMO-1 as indicated. Reactions were terminated by the addition of SDS loading buffer. Proteins were analysed by Western blotting using anti-GST antibody. (D) Cbx4 enhances SUMOylation of Dnmt3a in vitro. Purified GST–Dnmt3aN was incubated with 250 ng of E1 (Aos1/Uba2 heterodimer), 20 ng of E2 (Ubc9) and 3 μg of SUMO-1. Lanes 1–4 contained 0, 25, 50 or 100 ng of purified His-Cbx4 respectively. Note that the amount of Ubc9 used here was 5-fold lower than that used in (C).
Similar articles
- Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription.
Ling Y, Sankpal UT, Robertson AK, McNally JG, Karpova T, Robertson KD. Ling Y, et al. Nucleic Acids Res. 2004 Jan 29;32(2):598-610. doi: 10.1093/nar/gkh195. Print 2004. Nucleic Acids Res. 2004. PMID: 14752048 Free PMC article. - Dnmt3b, de novo DNA methyltransferase, interacts with SUMO-1 and Ubc9 through its N-terminal region and is subject to modification by SUMO-1.
Kang ES, Park CW, Chung JH. Kang ES, et al. Biochem Biophys Res Commun. 2001 Dec 14;289(4):862-8. doi: 10.1006/bbrc.2001.6057. Biochem Biophys Res Commun. 2001. PMID: 11735126 - A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity.
Merrill JC, Melhuish TA, Kagey MH, Yang SH, Sharrocks AD, Wotton D. Merrill JC, et al. PLoS One. 2010 Jan 20;5(1):e8794. doi: 10.1371/journal.pone.0008794. PLoS One. 2010. PMID: 20098713 Free PMC article. - Pc2 and SUMOylation.
Wotton D, Merrill JC. Wotton D, et al. Biochem Soc Trans. 2007 Dec;35(Pt 6):1401-4. doi: 10.1042/BST0351401. Biochem Soc Trans. 2007. PMID: 18031231 Review. - SUMO-targeted ubiquitin ligases.
Sriramachandran AM, Dohmen RJ. Sriramachandran AM, et al. Biochim Biophys Acta. 2014 Jan;1843(1):75-85. doi: 10.1016/j.bbamcr.2013.08.022. Epub 2013 Sep 7. Biochim Biophys Acta. 2014. PMID: 24018209 Review.
Cited by
- CBX4 suppresses CD8+ T cell antitumor immunity by reprogramming glycolytic metabolism.
Wang J, Jia W, Zhou X, Ma Z, Liu J, Lan P. Wang J, et al. Theranostics. 2024 Jun 17;14(10):3793-3809. doi: 10.7150/thno.95748. eCollection 2024. Theranostics. 2024. PMID: 38994031 Free PMC article. - Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium.
Mardaryev AN, Liu B, Rapisarda V, Poterlowicz K, Malashchuk I, Rudolf J, Sharov AA, Jahoda CA, Fessing MY, Benitah SA, Xu GL, Botchkarev VA. Mardaryev AN, et al. J Cell Biol. 2016 Jan 4;212(1):77-89. doi: 10.1083/jcb.201506065. Epub 2015 Dec 28. J Cell Biol. 2016. PMID: 26711500 Free PMC article. - CBX4 promotes antitumor immunity by suppressing Pdcd1 expression in T cells.
Ren L, Li Z, Zhou Y, Zhang J, Zhao Z, Wu Z, Zhao Y, Ju Y, Pang X, Sun X, Wang W, Zhang Y. Ren L, et al. Mol Oncol. 2023 Dec;17(12):2694-2708. doi: 10.1002/1878-0261.13516. Epub 2023 Oct 9. Mol Oncol. 2023. PMID: 37691307 Free PMC article. - Cutting Edge: Polycomb Repressive Complex 1 Subunit Cbx4 Positively Regulates Effector Responses in CD8 T Cells.
Melo GA, Xu T, Calôba C, Schutte A, Passos TO, Neto MAN, Brum G, Oliveira-Vieira B, Higa L, Monteiro FLL, Berbert L, Gonçalves ANA, Tanuri A, Viola JPB, Werneck MBF, Nakaya HI, Pipkin ME, Martinez GJ, Pereira RM. Melo GA, et al. J Immunol. 2023 Sep 1;211(5):721-726. doi: 10.4049/jimmunol.2200757. J Immunol. 2023. PMID: 37486206 Free PMC article. - Human polycomb 2 protein is a SUMO E3 ligase and alleviates substrate-induced inhibition of cystathionine beta-synthase sumoylation.
Agrawal N, Banerjee R. Agrawal N, et al. PLoS One. 2008;3(12):e4032. doi: 10.1371/journal.pone.0004032. Epub 2008 Dec 24. PLoS One. 2008. PMID: 19107218 Free PMC article.
References
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
- Bestor T. H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. - PubMed
- Okano M., Bell D. W., Haber D. A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. - PubMed
- Ge Y. Z., Pu M. T., Gowher H., Wu H. P., Ding J. P., Jeltsch A., Xu G. L. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 2004;279:25447–25454. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases