DNA modification by sulfur: analysis of the sequence recognition specificity surrounding the modification sites - PubMed (original) (raw)
DNA modification by sulfur: analysis of the sequence recognition specificity surrounding the modification sites
Jingdan Liang et al. Nucleic Acids Res. 2007.
Abstract
The Dnd (DNA degradation) phenotype, reflecting a novel DNA modification by sulfur in Streptomyces lividans 1326, was strongly aggravated when one (dndB) of the five genes (dndABCDE) controlling it was mutated. Electrophoretic banding patterns of a plasmid (pHZ209), reflecting DNA degradation, displayed a clear change from a preferential modification site in strain 1326 to more random modifications in the mutant. Fourteen randomly modifiable sites on pHZ209 were localized, and each seemed to be able to be modified only once. Residues in a region (5'-c-cGGCCgccg-3') including a highly conserved 4-bp central core (5'-GGCC-3') in a well-documented preferential modification site were assessed for their necessity by site-directed mutagenesis. While the central core (GGCC) was found to be stringently required in 1326 and in the mutant, 'gccg' flanking its right could either abolish or reduce the modification frequency only in the mutant, and two separate nucleotides to the left had no dramatic effect. The lack of essentiality of DndB for S-modification suggests that it might only be required for enhancing or stabilizing the activity of a protein complex at the required preferential modification site, or resolving secondary structures flanking the modifiable site(s), known to constitute an obstacle for efficient modification.
Figures
Figure 1.
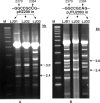
(A) Organization of the dnd gene cluster and in-frame deletion of _dnd_B. (B) Enhanced Dnd phenotype displayed by _dnd_B− mutant HXY2 as compared with that of wild-type 1326. Lack of Dnd phenotype displayed by _dnd_A− mutant HXY1 and _dnd_-deletion mutant HXY6 is shown as controls. (C) Preferential cleavage at specific site of EcoRV-linearized pHZ209 originating from 1326, but more random and lack of cleavage of the same DNA originating from HXY2 and HXY6, respectively. All the samples were treated with activated Tris-buffer followed by gel electrophoresis. The designation of the cleaved fragments conforms to those schematically outlined in Figure 3, with their sizes shown. DNA size markers are labeled ‘M’. Lagging of the DNA fragments behind the DNA marker could be seen as a result of overloading of the DNA samples.
Figure 2.
Restoration of modification preference towards EcoRV-linearized pHZ209 (A) and partial restoration of modification preference towards EcoRV-linearized pJTU2003 (in which the nucleotide ‘C’ at position 2384 of pHZ209 was mutated to ‘A’ (B), whose DNA came from LJD1 (with integration of the _dnd_B gene) and LJD2 (with integration of the complete dnd gene cluster), in contrast to the lack of restoration originating from LJD3 (with integration of the vector pSET152 carrying no dnd gene). DNA size markers are labeled ‘M’.
Figure 3.
(A) Alignment of the region traversing the modified sites (labeled as asterisks) sequenced by using pBluescript II SK(+) (left) or pMD18-T (right) as vectors for the identification of the central four nucleotides (GGCC) as the highly consensus region, which lies within one (DR2, underlining the corresponding sequence) of the three direct repeat sequences (DR1-3) in a region with a preferentially modified site (5L/R), enlarged to show the details of the region possessing three direct repeats (DR1-3, solid arrows) and two pairs of inverted repeats (IR1-2, framed arrows). Sequence tags conform to the relative positions in Figure 1C and in the linearized pHZ209 schematically presented at the bottom (B), while those labeled L or R are G site at either 2379 or 2380 in the sequenced terminus.
Figure 4.
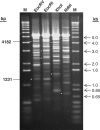
Banding patterns of pHZ2031 respectively linearized using four different but unique restriction enzymes (EcoRV, EcoRI, XhoI and NdeI) before treatment with activated Tris-buffer. Two fragments adding up to the size of linearized pHZ2031 (5.4 kb) were always seen at their expected positions after cleavage at a common modification site, as exemplified by cleavage at specific modification site 10 (Figure 3), indicated as two white asterisks in each respective gel panel. (EcoRV: 4182 and 1231; EcoRI: 3027 and 2386; XhoI: 3976 and 1437; NdeI: 4687 and 726).
Figure 5.
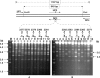
Site-directed mutagenesis of a region traversing the consensus sequence by a two-step PCR technique involving use of two variable oligonucleotide primers (DP2 and UP3) for introducing mutations at variable positions (indicated as a star in the primer region) with two fixed primers at both ends. The 949-bp DNA with site-specific mutations (all changed to A, see text and Table 2) at variable sites (specific nucleotide numbers indicated above each gel lane) could be obtained to replace the corresponding region on pHZ209 after digestion of the respective 1187-bp PCR products with EcoRI and XhoI. The individual constructs with desired replacements (confirmed by sequencing) were introduced into 1326 (heads with no dots) and HXY2 (heads with dots) before their respective DNA samples were isolated for assay in gel A and B. M stands for markers whose sizes are shown at the left of the gel.
Similar articles
- Twenty years hunting for sulfur in DNA.
Chen S, Wang L, Deng Z. Chen S, et al. Protein Cell. 2010 Jan;1(1):14-21. doi: 10.1007/s13238-010-0009-y. Epub 2010 Feb 7. Protein Cell. 2010. PMID: 21203994 Free PMC article. Review. - A novel DNA modification by sulphur.
Zhou X, He X, Liang J, Li A, Xu T, Kieser T, Helmann JD, Deng Z. Zhou X, et al. Mol Microbiol. 2005 Sep;57(5):1428-38. doi: 10.1111/j.1365-2958.2005.04764.x. Mol Microbiol. 2005. PMID: 16102010 - A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans.
You D, Wang L, Yao F, Zhou X, Deng Z. You D, et al. Biochemistry. 2007 May 22;46(20):6126-33. doi: 10.1021/bi602615k. Epub 2007 May 1. Biochemistry. 2007. PMID: 17469805 - Phosphorothioation of DNA in bacteria by dnd genes.
Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, Zhou X, You D, Deng Z, Dedon PC. Wang L, et al. Nat Chem Biol. 2007 Nov;3(11):709-10. doi: 10.1038/nchembio.2007.39. Epub 2007 Oct 14. Nat Chem Biol. 2007. PMID: 17934475 - DNA phosphorothioation in Streptomyces lividans: mutational analysis of the dnd locus.
Xu T, Liang J, Chen S, Wang L, He X, You D, Wang Z, Li A, Xu Z, Zhou X, Deng Z. Xu T, et al. BMC Microbiol. 2009 Feb 20;9:41. doi: 10.1186/1471-2180-9-41. BMC Microbiol. 2009. PMID: 19232083 Free PMC article.
Cited by
- Twenty years hunting for sulfur in DNA.
Chen S, Wang L, Deng Z. Chen S, et al. Protein Cell. 2010 Jan;1(1):14-21. doi: 10.1007/s13238-010-0009-y. Epub 2010 Feb 7. Protein Cell. 2010. PMID: 21203994 Free PMC article. Review. - Phosphorothioated DNA Is Shielded from Oxidative Damage.
Pu T, Liang J, Mei Z, Yang Y, Wang J, Zhang W, Liang WJ, Zhou X, Deng Z, Wang Z. Pu T, et al. Appl Environ Microbiol. 2019 Apr 4;85(8):e00104-19. doi: 10.1128/AEM.00104-19. Print 2019 Apr 15. Appl Environ Microbiol. 2019. PMID: 30737351 Free PMC article. - Purification, crystallization and preliminary X-ray analysis of the DndE protein from Salmonella enterica serovar Cerro 87, which is involved in DNA phosphorothioation.
Chen F, Lin K, Zhang Z, Chen L, Shi X, Cao C, Wang Z, Liang J, Deng Z, Wu G. Chen F, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011 Nov 1;67(Pt 11):1440-2. doi: 10.1107/S1744309111036694. Epub 2011 Oct 27. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011. PMID: 22102252 Free PMC article. - Phosphorothioate DNA as an antioxidant in bacteria.
Xie X, Liang J, Pu T, Xu F, Yao F, Yang Y, Zhao YL, You D, Zhou X, Deng Z, Wang Z. Xie X, et al. Nucleic Acids Res. 2012 Oct;40(18):9115-24. doi: 10.1093/nar/gks650. Epub 2012 Jul 5. Nucleic Acids Res. 2012. PMID: 22772986 Free PMC article. - Crystallization and preliminary X-ray analysis of the type IV restriction endonuclease ScoMcrA from Streptomyces coelicolor, which cleaves both Dcm-methylated DNA and phosphorothioated DNA.
Liu G, Zhang Z, Zhao G, Deng Z, Wu G, He X. Liu G, et al. Acta Crystallogr F Struct Biol Commun. 2015 Jan 1;71(Pt 1):57-60. doi: 10.1107/S2053230X14025801. Epub 2015 Jan 1. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 25615970 Free PMC article.
References
- Evans M., Kaczmarek F.S., Stutzman-Engwall K., Dyson P. Characterization of a Streptomyces-lividans-type site-specific DNA modification system in the avermectin-producer Streptomyces avermitilis permits investigation of two novel giant linear plasmids, pSA1 and pSA2. Microbiology. 1994;140(Pt. 6):1367–1371. - PubMed
- Ray T., Weaden J., Dyson P. Tris-dependent site-specific cleavage of Streptomyces lividans DNA. FEMS Microbiol. Lett. 1992;75:247–252. - PubMed
- Zhou X.F., He X.Y., Liang J.D., Li A.Y., Xu T.G., Kieser T., Helmann J.D., Deng Z.X. A novel DNA modification by sulphur. Mol. Microbiol. 2005;57:1428–1438. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases