UbiProt: a database of ubiquitylated proteins - PubMed (original) (raw)
UbiProt: a database of ubiquitylated proteins
Alexander L Chernorudskiy et al. BMC Bioinformatics. 2007.
Abstract
Background: Post-translational protein modification with ubiquitin, or ubiquitylation, is one of the hottest topics in a modern biology due to a dramatic impact on diverse metabolic pathways and involvement in pathogenesis of severe human diseases. A great number of eukaryotic proteins was found to be ubiquitylated. However, data about particular ubiquitylated proteins are rather disembodied.
Description: To fill a general need for collecting and systematizing experimental data concerning ubiquitylation we have developed a new resource, UbiProt Database, a knowledge base of ubiquitylated proteins. The database contains retrievable information about overall characteristics of a particular protein, ubiquitylation features, related ubiquitylation and de-ubiquitylation machinery and literature references reflecting experimental evidence of ubiquitylation. UbiProt is available at http://ubiprot.org.ru for free.
Conclusion: UbiProt Database is a public resource offering comprehensive information on ubiquitylated proteins. The resource can serve as a general reference source both for researchers in ubiquitin field and those who deal with particular ubiquitylated proteins which are of their interest. Further development of the UbiProt Database is expected to be of common interest for research groups involved in studies of the ubiquitin system.
Figures
Figure 1
Structure of a typical UbiProt entry. This entry contains information about an ubiquitylated NEMO protein.
Figure 2
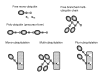
Different forms of ubiquitin and ubiquitin-modified proteins. Ubiquitylation may result in addition of a single ubiquitin moiety or a branched multi-ubiquitin chain to the target protein lysine(s). Note that a ubiquitin molecule possesses 7 inner lysine residues that can serve as attachment sites of the next ubiquitin moiety, resulting in the formation of the chains that have a different structure and topology. Functionally significant amino acids are marked as follows: Kn and Kn' – lysine residue(s) that can serve as attachment sites of the ubiquitin moiety; G76 – ubiquitin C-terminal glycine residue participating in the isopeptide bond formation. Poly-ubiquitin refers to a ubiquitin fusion protein, a precursor form polymerized "head-to-tail".
Figure 3
Example of full-text search results. The search pattern is "ubch5*" that corresponds to the particular E2 ubiquitin-conjugation enzyme (UbcH5). The result shows proteins to be modified by the specified enzyme.
Similar articles
- ProCMD: a database and 3D web resource for protein C mutants.
D'Ursi P, Marino F, Caprera A, Milanesi L, Faioni EM, Rovida E. D'Ursi P, et al. BMC Bioinformatics. 2007 Mar 8;8 Suppl 1(Suppl 1):S11. doi: 10.1186/1471-2105-8-S1-S11. BMC Bioinformatics. 2007. PMID: 17430555 Free PMC article. - Protein structure databases with new web services for structural biology and biomedical research.
Standley DM, Kinjo AR, Kinoshita K, Nakamura H. Standley DM, et al. Brief Bioinform. 2008 Jul;9(4):276-85. doi: 10.1093/bib/bbn015. Epub 2008 Apr 22. Brief Bioinform. 2008. PMID: 18430752 Review. - Proteomics FASTA archive and reference resource.
Falkner JA, Hill JA, Andrews PC. Falkner JA, et al. Proteomics. 2008 May;8(9):1756-7. doi: 10.1002/pmic.200701194. Proteomics. 2008. PMID: 18442177 - MtbSD--a comprehensive structural database for Mycobacterium tuberculosis.
Hassan S, Logambiga P, Raman AM, Subazini TK, Kumaraswami V, Hanna LE. Hassan S, et al. Tuberculosis (Edinb). 2011 Nov;91(6):556-62. doi: 10.1016/j.tube.2011.08.003. Epub 2011 Aug 30. Tuberculosis (Edinb). 2011. PMID: 21880546 - The Pain Genes Database: An interactive web browser of pain-related transgenic knockout studies.
Lacroix-Fralish ML, Ledoux JB, Mogil JS. Lacroix-Fralish ML, et al. Pain. 2007 Sep;131(1-2):3.e1-4. doi: 10.1016/j.pain.2007.04.041. Epub 2007 Jun 14. Pain. 2007. PMID: 17574758 Review.
Cited by
- The E3-ubiquitin ligase TRIM50 interacts with HDAC6 and p62, and promotes the sequestration and clearance of ubiquitinated proteins into the aggresome.
Fusco C, Micale L, Egorov M, Monti M, D'Addetta EV, Augello B, Cozzolino F, Calcagnì A, Fontana A, Polishchuk RS, Didelot G, Reymond A, Pucci P, Merla G. Fusco C, et al. PLoS One. 2012;7(7):e40440. doi: 10.1371/journal.pone.0040440. Epub 2012 Jul 9. PLoS One. 2012. PMID: 22792322 Free PMC article. - PupDB: a database of pupylated proteins.
Tung CW. Tung CW. BMC Bioinformatics. 2012 Mar 16;13:40. doi: 10.1186/1471-2105-13-40. BMC Bioinformatics. 2012. PMID: 22424087 Free PMC article. - E3Miner: a text mining tool for ubiquitin-protein ligases.
Lee H, Yi GS, Park JC. Lee H, et al. Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W416-22. doi: 10.1093/nar/gkn286. Epub 2008 May 15. Nucleic Acids Res. 2008. PMID: 18483079 Free PMC article. - DbPTM 3.0: an informative resource for investigating substrate site specificity and functional association of protein post-translational modifications.
Lu CT, Huang KY, Su MG, Lee TY, Bretaña NA, Chang WC, Chen YJ, Chen YJ, Huang HD. Lu CT, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D295-305. doi: 10.1093/nar/gks1229. Epub 2012 Nov 27. Nucleic Acids Res. 2013. PMID: 23193290 Free PMC article. - hUbiquitome: a database of experimentally verified ubiquitination cascades in humans.
Du Y, Xu N, Lu M, Li T. Du Y, et al. Database (Oxford). 2011 Nov 30;2011:bar055. doi: 10.1093/database/bar055. Print 2011. Database (Oxford). 2011. PMID: 22134927 Free PMC article.
References
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources