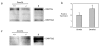Coordinate regulation of DNA methyltransferase expression during oogenesis - PubMed (original) (raw)
Comparative Study
Coordinate regulation of DNA methyltransferase expression during oogenesis
Diana Lucifero et al. BMC Dev Biol. 2007.
Abstract
Background: Normal mammalian development requires the action of DNA methyltransferases (DNMTs) for the establishment and maintenance of DNA methylation within repeat elements and imprinted genes. Here we report the expression dynamics of Dnmt3a and Dnmt3b, as well as a regulator of DNA methylation, Dnmt3L, in isolated female germ cells.
Results: Our results indicate that these enzymes are coordinately regulated and that their expression peaks during the stage of postnatal oocyte development when maternal methylation imprints are established. We find that Dnmt3a, Dnmt3b, Dnmt3L and Dnmt1o transcript accumulation is related to oocyte diameter. Furthermore, DNMT3L deficient 15 dpp oocytes have aberrantly methylated Snrpn, Peg3 and Igf2r DMRs, but normal IAP and LINE-1 methylation levels, thereby highlighting a male germ cell specific role for DNMT3L in the establishment of DNA methylation at repeat elements. Finally, real-time RT-PCR analysis indicates that the depletion of either DNMT3L or DNMT1o in growing oocytes results in the increased expression of the de novo methyltransferase Dnmt3b, suggesting a potential compensation mechanism by this enzyme for the loss of one of the other DNA methyltransferases.
Conclusion: Together these results provide a better understanding of the developmental regulation of Dnmt3a, Dnmt3b and Dnmt3L at the time of de novo methylation during oogenesis and demonstrate that the involvement of DNMT3L in retrotransposon silencing is restricted to the male germ line. This in turn suggests the existence of other factors in the oocyte that direct DNA methylation to transposons.
Figures
Figure 1
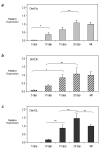
Developmental expression profiles of Dnmt3a, Dnmt3b, and Dnmt3L during postnatal oogenesis. QRT-PCR was used to determine the expression profile of a) Dnmt3a (light grey bars) b) Dnmt3b (cross hatch bars) and c) Dnmt3L (dark grey bars) in postnatal oocytes. Relative expression values obtained were normalized to the level of rabbit α-globin expression for each sample and were calibrated to the MII oocyte expression value. Representative results from one of the two independent experiments performed are plotted (see Additional file 2 for the second replicate). While the relative abundance of the transcripts varied for each of the timepoints, overall Dnmt3a, Dnmt3b and Dnmt3L expression increased with oocyte growth and peaked in 25 dpp oocytes for each enzyme. Results are presented as mean ± SD. * indicates significance of p < 0.05; ** indicates significance of p < 0.01.
Figure 2
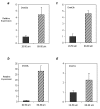
The effect of oocyte diameter on a) Dnmt3a, b) Dnmt3b, c) Dnmt3L and d) Dnmt1o expression. Oocytes were isolated at 10 dpp and pooled according to whether they were 20 to 50 μm or 60 to 80 μm in diameter. QRT-PCR was used to determine the relative expression of each of the enzymes in "small" (dark grey bars) vs. "big" (cross hatch bars) 10 dpp oocytes. One experiment where samples were tested in triplicate was carried out and relative expression values obtained were normalized to the level of rabbit α-globin expression for each sample and calibrated to the expression in 20 to 50 μm oocytes. For each of the enzymes investigated, the relative expression in "big" oocytes was at least 2- and up to 28-fold higher than that in "small" oocytes. Results are presented as mean ± SD. Differences between the relative expression in 20 to 50 μm and 60 to 80 μm oocytes were found to be statistically significant for each of the genes (p < 0.05).
Figure 3
Methylation profile of a) Snrpn, Peg3, Igf2r, H19 and b,c) IAP and LINE-1 repeats in _Dnmt3L_-/- oocytes. a,b) Bisulfite sequencing results are depicted as follows: each line represents an individual clone; a filled circle indicates a methylated CpG site, an open circle denotes an unmethylated CpG and a missing circle represents a CpG site where the sequencing data were ambiguous. H19 methylation served as control for somatic cell contamination. While Snrpn, Peg3 and Igf2r appear to be abnormally unmethylated (a), IAP LTR methylation is unaffected in Dnmt3L homozygous oocytes at 15 dpp (b). c) Methylation sensitive Southern analysis of IAP LTR and LINE-1 5'UTR methylation in Dnmt3L +/+ and -/- cumulus cells (soma) and mature metaphase II oocytes (oo). Lanes headed by M and H contain DNA that was cleaved with MspI and HpaII, respectively. Repeat sequences appear to be normally methylated in DNMT3L depleted oocytes.
Figure 4
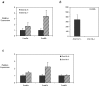
Dnmt3a, Dnmt3b and Dnmt3L expression in DNMT3L deficient 15 dpp oocytes and in DNMT1o deficient 25 dpp GV oocytes. a) QRT-PCR was used to analyze the expression of the de novo DNMT enzymes in Dnmt3L heterozygous (dark grey bars) and homozygous (cross hatch bars) oocytes at 15 dpp. Results for Dnmt3a and Dnmt3b are shown in a) while relative expression of Dnmt3L is illustrated in b). Dnmt3a and Dnmt3b transcripts are up-regulated in DNMT3L depleted growing oocytes. The differences observed for Dnmt3b and Dnmt3L were statistically significant (p < 0.01). c) QRT-PCR was used to analyze the expression of the DNMT enzymes in Dnmt1o heterozygous (dark grey bars) and homozygous (cross hatch bars) 25 dpp GV stage oocytes. While the relative expression of Dnmt3a and Dnmt3b was up-regulated in DNMT1o depleted GV oocytes, Dnmt3L expression remained unchanged. Samples were analyzed in triplicate and relative expression values obtained were normalized to the level of rabbit α-globin expression for each sample and were calibrated to the expression in heterozygous oocytes for Dnmt3a, Dnmt3b and Dnmt3L. For Dnmt3L analysis, expression was calibrated to the expression in homozygous oocytes. Results for one experiment are presented as mean ± SD.
Figure 5
Protein and transcript variant analysis of Dnmt3a and Dnmt3b in DNMT3L wild type and depleted 15 dpp oocytes. Western Blot analysis of a) DNMT3A and c) DNMT3B was carried out on growing oocytes isolated from Dnmt3L wild-type and homozygous 15 dpp females (280 oocytes/lane). For comparison and isoform identification, the panels on the right labelled 'B' show blotting results for type B spermatogonia (for the loading control see Additional file 4). As suggested by the RNA expression analysis in (b), both DNMT3a and DNMT3a2 are present in growing oocytes and the ratio of these transcripts does not change significantly in Dnmt3L homozygous oocytes. DNMT3b2 is the more abundant isoform in wild-type Dnmt3L oocytes and appears to be upregulated in mutant oocytes as also suggested by the RNA expression data in Figure 4a. b) Relative expression of Dnmt3a and Dnmt3a2 was determined using transcript specific primers. Expression was normalized to 18S. As previously mentioned, both transcripts are expressed in growing oocytes.
Figure 6
Onset of DNA methyltransferase expression during mammalian germ cell development. The results presented in this paper as well as observations made for the male germ line [25, 26] are depicted here in relationship to the timing of de novo methylation establishment on imprinted genes and repeats during germ cell development. The figure is adapted from [47]. The top panel illustrates the methylation dynamics of maternally and paternally methylated imprinted genes, depicted by the red and blue lines respectively. During gametogenesis the pattern of non-imprinted gene methylation closely resembles that of imprinted genes. The time at which DNA methyltransferases are expressed during germ cell development are indicated with the transcripts examined in this study highlighted in bold. Although expressed at similar stages of oogenesis, the de novo DNA methyltransferases, Dnmt3a and Dnmt3b, are grouped separately from Dnmt3L which does not have DNA methyltransferase activity. The onset of Dnmt1o expression has been previously shown in early growing oocytes [43]. Our findings show that the expression of Dnmt3a, Dnmt3b and Dnmt3L coincide with the establishment of DNA methylation on imprinted genes and repeat sequences in the female germ line. The progression of methylation imprint acquisition during oogenesis is illustrated in the bottom panel and depicted by the red shading above the female germ cells. PGC = primordial germ cell, Oog = oogonia, PL = preleptotene, L = leptotene, Z = zygotene, P = pachytene, NG = non-growing oocyte G = growing oocyte, MII = metaphase II oocyte.
Similar articles
- Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse.
Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. Kato Y, et al. Hum Mol Genet. 2007 Oct 1;16(19):2272-80. doi: 10.1093/hmg/ddm179. Epub 2007 Jul 6. Hum Mol Genet. 2007. PMID: 17616512 - Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells.
La Salle S, Mertineit C, Taketo T, Moens PB, Bestor TH, Trasler JM. La Salle S, et al. Dev Biol. 2004 Apr 15;268(2):403-15. doi: 10.1016/j.ydbio.2003.12.031. Dev Biol. 2004. PMID: 15063176 - Bovine DNA methylation imprints are established in an oocyte size-specific manner, which are coordinated with the expression of the DNMT3 family proteins.
O'Doherty AM, O'Shea LC, Fair T. O'Doherty AM, et al. Biol Reprod. 2012 Mar 19;86(3):67. doi: 10.1095/biolreprod.111.094946. Print 2012 Mar. Biol Reprod. 2012. PMID: 22088914 - Regulation of expression and activity of DNA (cytosine-5) methyltransferases in mammalian cells.
Kinney SR, Pradhan S. Kinney SR, et al. Prog Mol Biol Transl Sci. 2011;101:311-33. doi: 10.1016/B978-0-12-387685-0.00009-3. Prog Mol Biol Transl Sci. 2011. PMID: 21507356 Review. - Sexual dimorphism in parental imprint ontogeny and contribution to embryonic development.
Bourc'his D, Proudhon C. Bourc'his D, et al. Mol Cell Endocrinol. 2008 Jan 30;282(1-2):87-94. doi: 10.1016/j.mce.2007.11.025. Epub 2007 Nov 29. Mol Cell Endocrinol. 2008. PMID: 18178305 Review.
Cited by
- Maternal loss-of-function of Nlrp2 results in failure of epigenetic reprogramming in mouse oocytes.
Anvar Z, Jochum MD, Chakchouk I, Sharif M, Demond H, To AK, Kraushaar DC, Wan YW, Andrews S, Kelsey G, Veyver IB. Anvar Z, et al. Res Sq [Preprint]. 2024 Jun 4:rs.3.rs-4457414. doi: 10.21203/rs.3.rs-4457414/v1. Res Sq. 2024. PMID: 38883732 Free PMC article. Preprint. - Oocyte Aging: A Multifactorial Phenomenon in A Unique Cell.
Kordowitzki P, Graczyk S, Haghani A, Klutstein M. Kordowitzki P, et al. Aging Dis. 2024 Feb 1;15(1):5-21. doi: 10.14336/AD.2023.0527. Aging Dis. 2024. PMID: 37307833 Free PMC article. Review. - Food Safety, Animal Health and Welfare and Environmental Impact of Animals derived from Cloning by Somatic Cell Nucleus Transfer (SCNT) and their Offspring and Products Obtained from those Animals.
European Food Safety Authority (EFSA). European Food Safety Authority (EFSA). EFSA J. 2008 Jul 24;6(7):767. doi: 10.2903/j.efsa.2008.767. eCollection 2008 Jul. EFSA J. 2008. PMID: 37213844 Free PMC article. No abstract available. - Transgenerational inheritance and its modulation by environmental cues.
Verdikt R, Armstrong AA, Allard P. Verdikt R, et al. Curr Top Dev Biol. 2023;152:31-76. doi: 10.1016/bs.ctdb.2022.10.002. Epub 2022 Nov 14. Curr Top Dev Biol. 2023. PMID: 36707214 Free PMC article. - Transient Polycomb activity represses developmental genes in growing oocytes.
Jarred EG, Qu Z, Tsai T, Oberin R, Petautschnig S, Bildsoe H, Pederson S, Zhang QH, Stringer JM, Carroll J, Gardner DK, Van den Buuse M, Sims NA, Gibson WT, Adelson DL, Western PS. Jarred EG, et al. Clin Epigenetics. 2022 Dec 21;14(1):183. doi: 10.1186/s13148-022-01400-w. Clin Epigenetics. 2022. PMID: 36544159 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous