Silencing by imprinted noncoding RNAs: is transcription the answer? - PubMed (original) (raw)
Review
Silencing by imprinted noncoding RNAs: is transcription the answer?
Florian M Pauler et al. Trends Genet. 2007 Jun.
Abstract
Non-coding RNAs (ncRNAs) with gene regulatory functions are starting to be seen as a common feature of mammalian gene regulation with the discovery that most of the transcriptome is ncRNA. The prototype has long been the Xist ncRNA, which induces X-chromosome inactivation in female cells. However, a new paradigm is emerging--the silencing of imprinted gene clusters by long ncRNAs. Here, we review models by which imprinted ncRNAs could function. We argue that an Xist-like model is only one of many possible solutions and that imprinted ncRNAs could provide the better model for understanding the function of the new class of ncRNAs associated with non-imprinted mammalian genes.
Figures
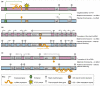
Figure 1
Genomic organization of three imprinted mouse clusters containing ncRNAs tested for their silencing function. Only a few genes in each cluster show widespread imprinted expression in extra-embryonic, embryonic and adult tissues, and most genes in a cluster show imprinted expression only in extra-embryonic tissues; thus, the expression pattern in this cell lineage is illustrated. (a) The paternally imprinted Igf2 cluster spans ~80 kb and contains the 2.5-kb H19 ncRNA [68]. Igf2 and H19 show ubiquitous imprinted expression, whereas Ins2 shows imprinted expression only in the extra-embryonic visceral yolk sac. Imprinted expression operates through the ICE, which is itself regulated by a DNA methylation imprint. On the maternal chromosome (pink), the CTCF protein binds to the unmethylated maternal ICE and forms an insulator that blocks the interaction of downstream enhancers to the Igf2 and Ins2 promoters but allows their interaction with the H19 ncRNA promoter. On the paternal chromosome (blue), the DNA methylation imprint blocks CTCF binding. Thus, the enhancers interact with the Igf2 and Ins2 promoters [66]. (b) The maternally imprinted Kcnq1 cluster spans ~800 kb and contains the Kcnq1ot1 ncRNA [16]. The Kcnq1ot1 promoter lies in an antisense orientation in the ICE in intron 10 of the Kcnq1 gene; its full length is unknown, as is whether it overlaps the Kcnq1 promoter. Cdkn1c and the Kcnq1ot1 ncRNA show ubiquitous imprinted expression, whereas the remainder show only limited imprinted expression in the early embryo or in extra-embryonic tissues. Imprinted expression operates through the ICE that contains the Kcnq1ot1 ncRNA promoter. On the paternal chromosome, the Kcnq1ot1 ncRNA is expressed and eight imprinted mRNA promoters are repressed. On the maternal chromosome, the methylated ICE represses the Kcnq1ot1 ncRNA promoter, enabling expression of the eight imprinted mRNAs only on this chromosome [4]. In this cluster, two genes (Nap1/4 and Phemx) show expression from both alleles, although expression of the paternal is less than that of the maternal allele [42]. (c) The maternally imprinted Igf2r cluster spans ~500 kb and contains the 108-kb Air ncRNA [67]. The Air promoter lies in antisense orientation in the ICE in Igf2r intron 2. In this cluster, Igf2r and the Air ncRNA show ubiquitous imprinted expression, whereas the remainder of the genes in the cluster show imprinted expression only in extra-embryonic tissues. Imprinted expression operates through the ICE that contains the Air ncRNA promoter. On the paternal chromosome, the Air ncRNA is expressed and three imprinted mRNA promoters are repressed. On the maternal chromosome, the methylated ICE represses the Air ncRNA promoter, enabling expression of the three imprinted mRNAs only on this chromosome [5]. Gene loci (not to scale) for which the gene name is not given in full are abbreviated as follows: Asc, Ascl2; Cd8, Cd81; Cdk, Cdkn1c; Nap, Nap1/4; Osb, Osbpl5; Phe, Phemx; Phl, Phlda2; Slc, Slc22a18; Trp, Trpm5; Tss, Tssc4.
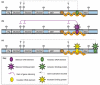
Figure 2
Gene-silencing models for the imprinted Igf2r cluster based on a role for the ncRNA product. Only the paternal expression pattern in extra-embryonic tissues is shown, in which the Air ncRNA is expressed and Igf2r, Slc22a2 and Slc22a3 are silenced. The remaining genes show tissue-specific silencing in this tissue. (a) RNAi-dependent silencing: the expressed ncRNA triggers RNAi by generating dsRNA, which subsequently induces transcriptional silencing at homologous regions in the cluster. dsRNA might arise either because of the transcriptional overlap with the ncRNA and a protein-coding gene that is found in both the Igf2r and Kcnq1 imprinted clusters, or from intrinsic properties of the ncRNA (i.e. the presence of inverted repeats), or from pairing with RNAs originating from interspersed repeats that are common to introns in all genes in the cluster. The Air ncRNA is unspliced and thus is rich in interspersed repeats. (b) RNA-directed targeting: the expressed ncRNA localizes to the chromosome that expresses it and can spread over the region containing silenced genes. Spreading could be limited by an absence of positive spreading signals on autosomes or be due to some type of insulator that blocks spreading. The ncRNA is subsequently able to recruit epigenetic silencing modifications in the form of DNA methylation, histone modifications or histone variants in a manner similar to that shown for the Xist ncRNA that mediates X-chromosome inactivation.
Figure 3
Transcription-based silencing models that do not require the ncRNA product itself, but only its transcription. (a) Transcription interference of _cis_-regulatory activator elements: ncRNA transcription displaces binding proteins from an activator that lies in the ncRNA gene body. This activator must be able to activate multiple genes, so is comparable to a locus control region (LCR) or could be considered a ‘domain’ activator. ncRNA transcription interferes with activator function by displacing binding proteins needed for long-range activation of all the imprinted protein-coding genes in the cluster. (b) Transcriptional activation of _cis_-regulatory silencing elements: ncRNA transcription activates a silencer (located in the ncRNA gene body) by enabling the binding of long-range silencer proteins that repress all imprinted protein-coding genes in the cluster. (c) Transcriptional activation of insulator elements: ncRNA transcription is proposed to activate an insulator or boundary element (located in the ncRNA gene body), which then enables binding of an insulator protein. Implicit in this model is that a domain activator needed for expression of all imprinted genes in the cluster (except the ncRNA) lies on the other side of the insulator element. Formation of the insulator prevents interaction between the domain activator and the imprinted genes.
Figure 4
Higher order chromatin silencing models. (a) Transcription-based looping model: ncRNA transcription and processing is proposed to induce looping of the DNA through interactions between its 5′ and 3′ processed ends. The induced chromatin loop physically isolates a domain activator needed for expression of all imprinted protein-coding genes in the cluster. In this model, the sequence of the ncRNA is not important for silencing function. Thus, this model would also fit the category of transcription-based models. (b) ncRNA-induced compartment model, based on the ncRNA forming a compartment that does not favour mRNA transcription. This could occur for many reasons: for example, the ncRNA physically excludes access to a transcription-compartment, or it could act as a sink to sequester RNA polymerase II, or it could create a subnuclear compartment that favours unspliced, non-exported RNAs.
Similar articles
- Epigenetics of imprinted long noncoding RNAs.
Mohammad F, Mondal T, Kanduri C. Mohammad F, et al. Epigenetics. 2009 Jul 1;4(5):277-86. Epub 2009 Jul 10. Epigenetics. 2009. PMID: 19617707 Review. - Non-coding RNAs in imprinted gene clusters.
Royo H, Cavaillé J. Royo H, et al. Biol Cell. 2008 Mar;100(3):149-66. doi: 10.1042/BC20070126. Biol Cell. 2008. PMID: 18271756 Review. - Long Noncoding RNAs and X Chromosome Inactivation.
Gontan C, Jonkers I, Gribnau J. Gontan C, et al. Prog Mol Subcell Biol. 2011;51:43-64. doi: 10.1007/978-3-642-16502-3_3. Prog Mol Subcell Biol. 2011. PMID: 21287133 - X chromosome inactivation sparked by non-coding RNAs.
Leeb M, Steffen PA, Wutz A. Leeb M, et al. RNA Biol. 2009 Apr-Jun;6(2):94-9. doi: 10.4161/rna.6.2.7716. Epub 2009 Apr 20. RNA Biol. 2009. PMID: 19246991 Review. - X inactivation: Tsix and Xist as yin and yang.
Mlynarczyk SK, Panning B. Mlynarczyk SK, et al. Curr Biol. 2000 Dec 14-28;10(24):R899-903. doi: 10.1016/s0960-9822(00)00847-2. Curr Biol. 2000. PMID: 11137025 Review.
Cited by
- Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm.
Zhang M, Zhao H, Xie S, Chen J, Xu Y, Wang K, Zhao H, Guan H, Hu X, Jiao Y, Song W, Lai J. Zhang M, et al. Proc Natl Acad Sci U S A. 2011 Dec 13;108(50):20042-7. doi: 10.1073/pnas.1112186108. Epub 2011 Nov 23. Proc Natl Acad Sci U S A. 2011. PMID: 22114195 Free PMC article. - Silencing by the imprinted Airn macro lncRNA: transcription is the answer.
Santoro F, Pauler FM. Santoro F, et al. Cell Cycle. 2013 Mar 1;12(5):711-2. doi: 10.4161/cc.23860. Epub 2013 Feb 19. Cell Cycle. 2013. PMID: 23422859 Free PMC article. No abstract available. - Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a.
Meng L, Person RE, Beaudet AL. Meng L, et al. Hum Mol Genet. 2012 Jul 1;21(13):3001-12. doi: 10.1093/hmg/dds130. Epub 2012 Apr 5. Hum Mol Genet. 2012. PMID: 22493002 Free PMC article. - Epigenetic mechanisms in mammals.
Kim JK, Samaranayake M, Pradhan S. Kim JK, et al. Cell Mol Life Sci. 2009 Feb;66(4):596-612. doi: 10.1007/s00018-008-8432-4. Cell Mol Life Sci. 2009. PMID: 18985277 Free PMC article. Review. - The emergence of lncRNAs in cancer biology.
Prensner JR, Chinnaiyan AM. Prensner JR, et al. Cancer Discov. 2011 Oct;1(5):391-407. doi: 10.1158/2159-8290.CD-11-0209. Cancer Discov. 2011. PMID: 22096659 Free PMC article. Review.
References
- Verona RI, et al. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 2003;19:237–259. - PubMed
- Barlow DP, Bartolomei MS. Genomic imprinting in mammals. In: Allis CD, et al., editors. Epigenetics. Cold Spring Harbor Laboratory Press; 2006. pp. 357–375.
- Lewis A, Reik W. How imprinting centres work. Cytogenet. Genome Res. 2006;113:81–89. - PubMed
- Regha K, et al. The imprinted mouse Igf2r/Air cluster – a model maternal imprinting system. Cytogenet. Genome Res. 2006;113:165–177. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources