Analysis of the first genome of a hyperthermophilic marine virus-like particle, PAV1, isolated from Pyrococcus abyssi - PubMed (original) (raw)
Analysis of the first genome of a hyperthermophilic marine virus-like particle, PAV1, isolated from Pyrococcus abyssi
C Geslin et al. J Bacteriol. 2007 Jun.
Abstract
Only one virus-like particle (VLP) has been reported from hyperthermophilic Euryarchaeotes. This VLP, named PAV1, is shaped like a lemon and was isolated from a strain of "Pyrococcus abyssi," a deep-sea isolate. Its genome consists of a double-stranded circular DNA of 18 kb which is also present at a high copy number (60 per chromosome) free within the host cytoplasm but is not integrated into the host chromosome. Here, we report the results of complete analysis of the PAV1 genome. All the 25 predicted genes, except 3, are located on one DNA strand. A transcription map has been made by using a reverse transcription-PCR assay. All the identified open reading frames (ORFs) are transcribed. The most significant similarities relate to four ORFs. ORF 180a shows 31% identity with ORF 181 of the pRT1 plasmid isolated from Pyrococcus sp. strain JT1. ORFs 676 and 678 present similarities with a concanavalin A-like lectin/glucanase domain, which could be involved in the process of host-virus recognition, and ORF 59 presents similarities with the transcriptional regulator CopG. The genome of PAV1 displays unique features at the nucleic and proteinic level, indicating that PAV1 should be attached at least to a novel genus or virus family.
Figures
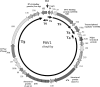
FIG. 1.
PAV1 genome map. Predicted genes are represented by thick arrows; light gray shading indicates ORFs with no similarity and no assigned function, and dark gray shading indicates either conserved hypothetical ORFs or ORFs with a hypothetical function; hatching indicates ORFs encoding a putative membrane-associated protein. Transcript locations mapped by using RT-PCR (T1 to T6) are shown by arrows inside the circular genome map. The approximate location of the origin (Ori) of replication as predicted by cumulative GC skew is also indicated. Protein motif or domain names: Lam G, laminin G; Leu zip, leucine zipper; P-loop, nucleoside triphosphate binding site or Walker motif A; wHTH, winged helix or winged helix-turn-helix.
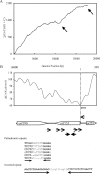
FIG. 2.
Structure of the putative DNA replication origin region. (A) Cumulative GC skew (window size of 20 bp). (B) Identification of a DNA unwinding element (DUE) and nucleotide repeats (indicated by arrows). The longer inverted repeats are indicated by the thick black arrows.
FIG. 3.
ORF 676 and ORF 678 possess domains related to laminin G-like jellyroll fold. Amino acid sequence alignment of the internal repeats of ORFs 676 and 678 with various members of the concanavalin A/glucanase structural family (human serum amyloid P component [1sac_A], hypothetical protein from bacteriophage S-PM2 [gi 58532986], VCBS from Pelodyction luteolum [gi 78186255], hypothetical protein from Rhodopirellula baltica [gi 32471540], VCBS from Prosthecochloris vibrioformis [gi 71481241], and S-layer protein from Clostridium thermocellum [gi 67915998]) are shown. The numbers in brackets indicate the positions in the sequences. The secondary elements of the human serum amyloid P component are displayed above the alignment.
Similar articles
- Crystal structure of PAV1-137: a protein from the virus PAV1 that infects Pyrococcus abyssi.
Leulliot N, Quevillon-Cheruel S, Graille M, Geslin C, Flament D, Le Romancer M, van Tilbeurgh H. Leulliot N, et al. Archaea. 2013;2013:568053. doi: 10.1155/2013/568053. Epub 2013 Mar 4. Archaea. 2013. PMID: 23533329 Free PMC article. - TPV1, the first virus isolated from the hyperthermophilic genus Thermococcus.
Gorlas A, Koonin EV, Bienvenu N, Prieur D, Geslin C. Gorlas A, et al. Environ Microbiol. 2012 Feb;14(2):503-16. doi: 10.1111/j.1462-2920.2011.02662.x. Epub 2011 Dec 12. Environ Microbiol. 2012. PMID: 22151304 Free PMC article. - PAV1, the first virus-like particle isolated from a hyperthermophilic euryarchaeote, "Pyrococcus abyssi".
Geslin C, Le Romancer M, Erauso G, Gaillard M, Perrot G, Prieur D. Geslin C, et al. J Bacteriol. 2003 Jul;185(13):3888-94. doi: 10.1128/JB.185.13.3888-3894.2003. J Bacteriol. 2003. PMID: 12813083 Free PMC article. - Diversity of CRISPR systems in the euryarchaeal Pyrococcales.
Norais C, Moisan A, Gaspin C, Clouet-d'Orval B. Norais C, et al. RNA Biol. 2013 May;10(5):659-70. doi: 10.4161/rna.23927. Epub 2013 Feb 19. RNA Biol. 2013. PMID: 23422322 Free PMC article. Review. - Exceptionally diverse morphotypes and genomes of crenarchaeal hyperthermophilic viruses.
Prangishvili D, Garrett RA. Prangishvili D, et al. Biochem Soc Trans. 2004 Apr;32(Pt 2):204-8. doi: 10.1042/bst0320204. Biochem Soc Trans. 2004. PMID: 15046572 Review.
Cited by
- Rising to the challenge: accelerated pace of discovery transforms marine virology.
Brum JR, Sullivan MB. Brum JR, et al. Nat Rev Microbiol. 2015 Mar;13(3):147-59. doi: 10.1038/nrmicro3404. Epub 2015 Feb 2. Nat Rev Microbiol. 2015. PMID: 25639680 Review. - Active virus-host interactions at sub-freezing temperatures in Arctic peat soil.
Trubl G, Kimbrel JA, Liquet-Gonzalez J, Nuccio EE, Weber PK, Pett-Ridge J, Jansson JK, Waldrop MP, Blazewicz SJ. Trubl G, et al. Microbiome. 2021 Oct 18;9(1):208. doi: 10.1186/s40168-021-01154-2. Microbiome. 2021. PMID: 34663463 Free PMC article. - A simple procedure to determine the infectivity and host range of viruses infecting anaerobic and hyperthermophilic microorganisms.
Gorlas A, Geslin C. Gorlas A, et al. Extremophiles. 2013 Mar;17(2):349-55. doi: 10.1007/s00792-013-0513-0. Epub 2013 Jan 23. Extremophiles. 2013. PMID: 23340763 - Living side by side with a virus: characterization of two novel plasmids from Thermococcus prieurii, a host for the spindle-shaped virus TPV1.
Gorlas A, Krupovic M, Forterre P, Geslin C. Gorlas A, et al. Appl Environ Microbiol. 2013 Jun;79(12):3822-8. doi: 10.1128/AEM.00525-13. Epub 2013 Apr 12. Appl Environ Microbiol. 2013. PMID: 23584787 Free PMC article. - Two novel families of plasmids from hyperthermophilic archaea encoding new families of replication proteins.
Soler N, Marguet E, Cortez D, Desnoues N, Keller J, van Tilbeurgh H, Sezonov G, Forterre P. Soler N, et al. Nucleic Acids Res. 2010 Aug;38(15):5088-104. doi: 10.1093/nar/gkq236. Epub 2010 Apr 18. Nucleic Acids Res. 2010. PMID: 20403814 Free PMC article.
References
- Acebo, P., M. Garcia de Lacoba, G. Rivas, J. M. Andreu, M. Espinosa, and G. del Solar. 1998. Structural features of the plasmid pMV158-encoded transcriptional repressor CopG, a protein sharing similarities with both helix-turn-helix and beta-sheet DNA binding proteins. Proteins 32:248-261. - PubMed
- Aquino, R. S., A. M. Landeira-Fernandez, A. P. Valente, L. R. Andrade, and P. A. Mourao. 2005. Occurrence of sulfated galactans in marine angiosperms: evolutionary implications. Glycobiology 15:11-20. - PubMed
- Arnold, H. P., U. Ziese, and W. Zillig. 2000. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology 272:409-416. - PubMed
- Arnold, H. P., W. Zillig, U. Ziese, I. Holz, M. Crosby, T. Utterback, J. F. Weidmann, J. K. Kristjansson, H. P. Klenk, K. E. Nelson, and C. M. Fraser. 2000. A novel lipothrixvirus, SIFV, of the extremely thermophilic crenarchaeon Sulfolobus. Virology 267:252-266. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources