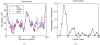vGNM: a better model for understanding the dynamics of proteins in crystals - PubMed (original) (raw)
vGNM: a better model for understanding the dynamics of proteins in crystals
Guang Song et al. J Mol Biol. 2007.
Abstract
The dynamics of proteins are important for understanding their functions. In recent years, the simple coarse-grained Gaussian Network Model (GNM) has been fairly successful in interpreting crystallographic B-factors. However, the model clearly ignores the contribution of the rigid body motions and the effect of crystal packing. The model cannot explain the fact that the same protein may have significantly different B-factors under different crystal packing conditions. In this work, we propose a new GNM, called vGNM, which takes into account both the contribution of the rigid body motions and the effect of crystal packing, by allowing the amplitude of the internal modes to be variables. It hypothesizes that the effect of crystal packing should cause some modes to be amplified and others to become less important. In doing so, vGNM is able to resolve the apparent discrepancy in experimental B-factors among structures of the same protein but with different crystal packing conditions, which GNM cannot explain. With a small number of parameters, vGNM is able to reproduce experimental B-factors for a large set of proteins with significantly better correlations (having a mean value of 0.81 as compared to 0.59 by GNM). The results of applying vGNM also show that the rigid body motions account for nearly 60% of the total fluctuations, in good agreement with previous findings.
Figures
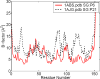
Figure 1
Example of two different myoglobin crystal structures showing different temperature factors. The two structures, 1ABS.pdb (space group: P6) and 1AJG.pdb (space group: P21), of the same protein (sperm whale myoglobin), display rather different B-factors. The correlation between the two is only 0.61.
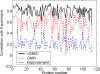
Figure 2
Comparison of the correlations between experimental and calculated B-factors for 113 proteins from GNM and vGNM. The improvement is shown in dot dash line. vGNM produces significantly better correlations with a mean value of 0.81 (over all the proteins) as compared to 0.59 from normal GNM.
Figure 3
(a) The B-factors calculated from vGNM and GNM for calmodulin (1osa.pdb). vGNM is able to reproduce extremely well the experimental B-factors. (b) the internal modes selected by vGNM, showing that the lowest frequency mode is not activated at all and the third lowest frequency mode makes the largest contribution.
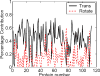
Figure 4
The percentage contributions of rigid body translation and rotation to the total fluctuations for all proteins. The mean rigid body contribution is about 59%, with 44% from translational and 15% from rotational motions.
Figure 5
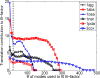
The translation contribution to B-factors (wtrans inEq. 7) varies as the number of low frequency modes that are included in the least squares fit increases, for six example proteins of varies sizes.The contribution usually becomes stabilized after a small number of modes are included. The vertical dashed line marks where the number of modes is 20.
Figure 6
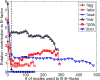
The magnitude of the rotation contribution to B-factors (wrotate in Eq. 7) varies as the number of low frequency modes that are included in the least squares fit increases, for six example proteins of varies sizes. The contribution becomes stabilized after a small number of modes are included. The vertical dashed line marks where the number of modes is 20.
Similar articles
- Dynamics of proteins in crystals: comparison of experiment with simple models.
Kundu S, Melton JS, Sorensen DC, Phillips GN Jr. Kundu S, et al. Biophys J. 2002 Aug;83(2):723-32. doi: 10.1016/S0006-3495(02)75203-X. Biophys J. 2002. PMID: 12124259 Free PMC article. - Generalized spring tensor models for protein fluctuation dynamics and conformation changes.
Na H, Lin TL, Song G. Na H, et al. Adv Exp Med Biol. 2014;805:107-35. doi: 10.1007/978-3-319-02970-2_5. Adv Exp Med Biol. 2014. PMID: 24446359 - oGNM: online computation of structural dynamics using the Gaussian Network Model.
Yang LW, Rader AJ, Liu X, Jursa CJ, Chen SC, Karimi HA, Bahar I. Yang LW, et al. Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W24-31. doi: 10.1093/nar/gkl084. Nucleic Acids Res. 2006. PMID: 16845002 Free PMC article. - Critical evaluation of simple network models of protein dynamics and their comparison with crystallographic B-factors.
Soheilifard R, Makarov DE, Rodin GJ. Soheilifard R, et al. Phys Biol. 2008 Jun 24;5(2):026008. doi: 10.1088/1478-3975/5/2/026008. Phys Biol. 2008. PMID: 18577808 - Comparing protein-ligand docking programs is difficult.
Cole JC, Murray CW, Nissink JW, Taylor RD, Taylor R. Cole JC, et al. Proteins. 2005 Aug 15;60(3):325-32. doi: 10.1002/prot.20497. Proteins. 2005. PMID: 15937897 Review.
Cited by
- Communication: Capturing protein multiscale thermal fluctuations.
Opron K, Xia K, Wei GW. Opron K, et al. J Chem Phys. 2015 Jun 7;142(21):211101. doi: 10.1063/1.4922045. J Chem Phys. 2015. PMID: 26049417 Free PMC article. - A comparative analysis of the equilibrium dynamics of a designed protein inferred from NMR, X-ray, and computations.
Liu L, Koharudin LM, Gronenborn AM, Bahar I. Liu L, et al. Proteins. 2009 Dec;77(4):927-39. doi: 10.1002/prot.22518. Proteins. 2009. PMID: 19688820 Free PMC article. - Sequence-based Gaussian network model for protein dynamics.
Zhang H, Kurgan L. Zhang H, et al. Bioinformatics. 2014 Feb 15;30(4):497-505. doi: 10.1093/bioinformatics/btt716. Epub 2013 Dec 12. Bioinformatics. 2014. PMID: 24336646 Free PMC article. - Fast and anisotropic flexibility-rigidity index for protein flexibility and fluctuation analysis.
Opron K, Xia K, Wei GW. Opron K, et al. J Chem Phys. 2014 Jun 21;140(23):234105. doi: 10.1063/1.4882258. J Chem Phys. 2014. PMID: 24952521 Free PMC article. - Flexibility-rigidity index for protein-nucleic acid flexibility and fluctuation analysis.
Opron K, Xia K, Burton Z, Wei GW. Opron K, et al. J Comput Chem. 2016 May 30;37(14):1283-95. doi: 10.1002/jcc.24320. Epub 2016 Mar 1. J Comput Chem. 2016. PMID: 26927815 Free PMC article.
References
- Song G, Jernigan RL. An enhanced elastic network model to represent the motions of domain-swapped proteins. Proteins. 2006;63:197–209. - PubMed
- Cruickshank DWJ. The analysis of the anisotropic thermal motion of molecules in crystals. Acta Cryst. 1956;9:754–756.
- Schomaker V, Trueblood KN. On the rigid-body motion of molecules in crystals. Acta Cryst. 1968;B24:63–76.
- Stec B, Zhou R, Teeter MM. Full-matrix refinement of the protein crambin at 0.83 a and 130 k. Acta Crystallogr D. 1995;51:663–81. - PubMed
- Winn MD, Isupov MN, Murshudov GN. Use of tls parameters to model anisotropic displacements in macromolecular refinement. Acta Cryst. 2001;D57:122–33. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources