OligoCalc: an online oligonucleotide properties calculator - PubMed (original) (raw)
. 2007 Jul;35(Web Server issue):W43-6.
doi: 10.1093/nar/gkm234. Epub 2007 Apr 22.
Affiliations
- PMID: 17452344
- PMCID: PMC1933198
- DOI: 10.1093/nar/gkm234
OligoCalc: an online oligonucleotide properties calculator
Warren A Kibbe. Nucleic Acids Res. 2007 Jul.
Abstract
We developed OligoCalc as a web-accessible, client-based computational engine for reporting DNA and RNA single-stranded and double-stranded properties, including molecular weight, solution concentration, melting temperature, estimated absorbance coefficients, inter-molecular self-complementarity estimation and intra-molecular hairpin loop formation. OligoCalc has a familiar 'calculator' look and feel, making it readily understandable and usable. OligoCalc incorporates three common methods for calculating oligonucleotide-melting temperatures, including a nearest-neighbor thermodynamic model for melting temperature. Since it first came online in 1997, there have been more than 900,000 accesses of OligoCalc from nearly 200,000 distinct hosts, excluding search engines. OligoCalc is available at http://basic.northwestern.edu/biotools/OligoCalc.html, with links to the full source code, usage patterns and statistics at that link as well.
Figures
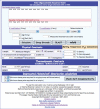
Figure 1.
Entry and main calculation screen for OligoCalc.
Similar articles
- dnaMATE: a consensus melting temperature prediction server for short DNA sequences.
Panjkovich A, Norambuena T, Melo F. Panjkovich A, et al. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W570-2. doi: 10.1093/nar/gki379. Nucleic Acids Res. 2005. PMID: 15980538 Free PMC article. - A fractional programming approach to efficient DNA melting temperature calculation.
Leber M, Kaderali L, Schönhuth A, Schrader R. Leber M, et al. Bioinformatics. 2005 May 15;21(10):2375-82. doi: 10.1093/bioinformatics/bti379. Epub 2005 Mar 15. Bioinformatics. 2005. PMID: 15769839 - INFO-RNA--a server for fast inverse RNA folding satisfying sequence constraints.
Busch A, Backofen R. Busch A, et al. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W310-3. doi: 10.1093/nar/gkm218. Epub 2007 Apr 22. Nucleic Acids Res. 2007. PMID: 17452349 Free PMC article. - Insights to primitive replication derived from structures of small oligonucleotides.
Smith GK, Fox GE. Smith GK, et al. Microbiologia. 1995;11:217-24. Microbiologia. 1995. PMID: 11539564 Review. - Computational approaches for investigating base flipping in oligonucleotides.
Priyakumar UD, MacKerell AD Jr. Priyakumar UD, et al. Chem Rev. 2006 Feb;106(2):489-505. doi: 10.1021/cr040475z. Chem Rev. 2006. PMID: 16464016 Review. No abstract available.
Cited by
- De-MetaST-BLAST: a tool for the validation of degenerate primer sets and data mining of publicly available metagenomes.
Gulvik CA, Effler TC, Wilhelm SW, Buchan A. Gulvik CA, et al. PLoS One. 2012;7(11):e50362. doi: 10.1371/journal.pone.0050362. Epub 2012 Nov 26. PLoS One. 2012. PMID: 23189198 Free PMC article. - Induced folding in RNA recognition by Arabidopsis thaliana DCL1.
Suarez IP, Burdisso P, Benoit MP, Boisbouvier J, Rasia RM. Suarez IP, et al. Nucleic Acids Res. 2015 Jul 27;43(13):6607-19. doi: 10.1093/nar/gkv627. Epub 2015 Jun 22. Nucleic Acids Res. 2015. PMID: 26101256 Free PMC article. - Variable-temperature size exclusion chromatography for the study of the structural changes in g-quadruplex.
Benabou S, Eritja R, Gargallo R. Benabou S, et al. ISRN Biochem. 2013 Nov 10;2013:631875. doi: 10.1155/2013/631875. eCollection 2013. ISRN Biochem. 2013. PMID: 25937962 Free PMC article. - A DNA microarray for the authentication of giant tiger prawn (Penaeus monodon) and whiteleg shrimp (Penaeus (Litopenaeus) vannamei): a proof-of-principle.
Kappel K, Fafińska J, Fischer M, Fritsche J. Kappel K, et al. Anal Bioanal Chem. 2021 Aug;413(19):4837-4846. doi: 10.1007/s00216-021-03440-2. Epub 2021 Jun 11. Anal Bioanal Chem. 2021. PMID: 34114084 Free PMC article. - Intraspecific Polymorphisms of Cytogenetic Markers Mapped on Chromosomes of Triticum polonicum L.
Kwiatek M, Majka M, Majka J, Belter J, Suchowilska E, Wachowska U, Wiwart M, Wiśniewska H. Kwiatek M, et al. PLoS One. 2016 Jul 8;11(7):e0158883. doi: 10.1371/journal.pone.0158883. eCollection 2016. PLoS One. 2016. PMID: 27391447 Free PMC article.
References
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. - PubMed
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001.
- Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 1962;5:109–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources