MolProbity: all-atom contacts and structure validation for proteins and nucleic acids - PubMed (original) (raw)
. 2007 Jul;35(Web Server issue):W375-83.
doi: 10.1093/nar/gkm216. Epub 2007 Apr 22.
Affiliations
- PMID: 17452350
- PMCID: PMC1933162
- DOI: 10.1093/nar/gkm216
MolProbity: all-atom contacts and structure validation for proteins and nucleic acids
Ian W Davis et al. Nucleic Acids Res. 2007 Jul.
Abstract
MolProbity is a general-purpose web server offering quality validation for 3D structures of proteins, nucleic acids and complexes. It provides detailed all-atom contact analysis of any steric problems within the molecules as well as updated dihedral-angle diagnostics, and it can calculate and display the H-bond and van der Waals contacts in the interfaces between components. An integral step in the process is the addition and full optimization of all hydrogen atoms, both polar and nonpolar. New analysis functions have been added for RNA, for interfaces, and for NMR ensembles. Additionally, both the web site and major component programs have been rewritten to improve speed, convenience, clarity and integration with other resources. MolProbity results are reported in multiple forms: as overall numeric scores, as lists or charts of local problems, as downloadable PDB and graphics files, and most notably as informative, manipulable 3D kinemage graphics shown online in the KiNG viewer. This service is available free to all users at http://molprobity.biochem.duke.edu.
Figures
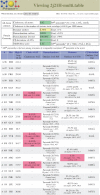
Figure 1.
Multi-criterion chart for 2J21, a crystal structure of the p53 DNA-binding core domain (37). The chart shows both overall statistics (top) and the first 20 residues of local data. Although a few steric clashes and one rotamer outlier are visible here (pink boxes) and might be worth trying to fix, this is an excellent structure overall; its resolution is 1.6Å, and compared to other structures at similar resolution, it ranks in the 92nd percentile for overall quality (MolProbity score).
Figure 2.
Multi-criterion kinemage of a 5-model NMR ensemble in side-by-side stereo, displayed in the KiNG applet. A small peptide (yellow) is bound to a short RNA hairpin (black), in file 1BIV (38). MolProbity has highlighted steric clashes (pink spikes), suspect RNA sugar puckers (magenta T's), outlier conformations of protein backbone (green) and sidechains (gold) and deviant bond angles around protein Cα's (magenta balls). On the right side, KiNG has controls for turning on or off the individual models, parts of the molecules (protein versus nucleic acid, Calphas versus full backbone, etc.) and validation criteria.
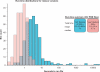
Figure 3.
Run-time distributions: improvement for the new version of Reduce. Optimizing hydrogens by exhaustive enumeration (cyan) was much slower than the new dynamic programming algorithm (pink). In many cases, this is the difference between a brief wait and completely infeasible calculation.
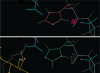
Figure 4.
Example of a flipped active-site histidine found and corrected by Reduce. Top: His 125 of the apo LpxA clashes (red spikes) with nearby Asp 126 rather than H-bonding (green dots), prompting Reduce to suggest flipping it. Bottom: Once the product (gold) was modeled into its electron density (32), it was evident that the flipped position is necessary for His 125 to participate in catalysis.
Similar articles
- MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes.
Davis IW, Murray LW, Richardson JS, Richardson DC. Davis IW, et al. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W615-9. doi: 10.1093/nar/gkh398. Nucleic Acids Res. 2004. PMID: 15215462 Free PMC article. - Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms.
Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, Richardson JS, Richardson DC. Word JM, et al. J Mol Biol. 1999 Jan 29;285(4):1711-33. doi: 10.1006/jmbi.1998.2400. J Mol Biol. 1999. PMID: 9917407 - WebFEATURE: An interactive web tool for identifying and visualizing functional sites on macromolecular structures.
Liang MP, Banatao DR, Klein TE, Brutlag DL, Altman RB. Liang MP, et al. Nucleic Acids Res. 2003 Jul 1;31(13):3324-7. doi: 10.1093/nar/gkg553. Nucleic Acids Res. 2003. PMID: 12824318 Free PMC article. - Modeling nucleic acids.
Sim AY, Minary P, Levitt M. Sim AY, et al. Curr Opin Struct Biol. 2012 Jun;22(3):273-8. doi: 10.1016/j.sbi.2012.03.012. Epub 2012 Apr 25. Curr Opin Struct Biol. 2012. PMID: 22538125 Free PMC article. Review. - New Biological Insights from Better Structure Models.
Touw WG, Joosten RP, Vriend G. Touw WG, et al. J Mol Biol. 2016 Mar 27;428(6):1375-1393. doi: 10.1016/j.jmb.2016.02.002. Epub 2016 Feb 8. J Mol Biol. 2016. PMID: 26869101 Review.
Cited by
- Crystal structure of the ubiquitin-like small archaeal modifier protein 2 from Haloferax volcanii.
Li Y, Maciejewski MW, Martin J, Jin K, Zhang Y, Maupin-Furlow JA, Hao B. Li Y, et al. Protein Sci. 2013 Sep;22(9):1206-17. doi: 10.1002/pro.2305. Epub 2013 Jul 27. Protein Sci. 2013. PMID: 23821306 Free PMC article. - The epitopes in wheat proteins for defining toxic units relevant to human health.
Juhász A, Gell G, Békés F, Balázs E. Juhász A, et al. Funct Integr Genomics. 2012 Nov;12(4):585-98. doi: 10.1007/s10142-012-0302-3. Epub 2012 Nov 20. Funct Integr Genomics. 2012. PMID: 23179564 Review. - Crystal structure of timothy grass allergen Phl p 12.0101 reveals an unusual profilin dimer.
O'Malley A, Kapingidza AB, Hyduke N, Dolamore C, Kowal K, Chruszcz M. O'Malley A, et al. Acta Biochim Pol. 2021 Mar 15;68(1):15-22. doi: 10.18388/abp.2020_5587. Acta Biochim Pol. 2021. PMID: 33720678 Free PMC article. - Interaction between DLC-1 and SAO-1 facilitates CED-4 translocation during apoptosis in the Caenorhabditis elegans germline.
Zhang D, Yang H, Jiang L, Zhao C, Wang M, Hu B, Yu C, Wei Z, Tse YC. Zhang D, et al. Cell Death Discov. 2022 Nov 3;8(1):441. doi: 10.1038/s41420-022-01233-9. Cell Death Discov. 2022. PMID: 36323675 Free PMC article. - The NAD+-mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Jiang Y, Liu T, Lee CH, Chang Q, Yang J, Zhang Z. Jiang Y, et al. Nature. 2020 Dec;588(7839):658-663. doi: 10.1038/s41586-020-2862-z. Epub 2020 Oct 14. Nature. 2020. PMID: 33053563
References
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. ProCheck - A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291.
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. - PubMed
- Vriend G. WHAT IF: A molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources