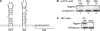RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation - PubMed (original) (raw)
RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation
J Robert Hogg et al. RNA. 2007 Jun.
Abstract
Recent studies have uncovered an unanticipated diversity of noncoding RNAs (ncRNAs), although these studies provide limited insight into their biological significance. Numerous general methods for identification and characterization of protein interactions have been developed, but similar approaches for characterizing cellular ncRNA interactions are lacking. Here we describe RNA Affinity in Tandem (RAT), an original, entirely RNA tag-based method for affinity purification of endogenously assembled RNP complexes. We demonstrate the general utility of RAT by isolating RNPs assembled in vivo on ncRNAs transcribed by RNA polymerase II or III. Using RAT in conjunction with protein identification by mass spectrometry and protein-RNA interaction assays, we define and characterize previously unanticipated protein subunits of endogenously assembled human 7SK RNPs. We show that 7SK RNA resides in a mixed population of RNPs with different protein compositions and responses to cellular stress. Depletion of a newly identified 7SK RNP component, hnRNP K, alters the partitioning of 7SK RNA among distinct RNPs. Our results establish the utility of a generalizable RNA-based RNP affinity purification method and provide insight into 7SK RNP dynamics.
Figures
FIGURE 1.
Expression of RAT-tagged 7SK RNAs. (A) RAT-7SK RNA. The 5′ end of human 7SK RNA was tagged with hairpins that bind PP7 coat protein (PP7) and tobramycin (Tob) as shown. (B) Various RAT-tagged 7SK RNAs were expressed using the human U6 RNA gene promoter subcloned into the vector backbone of pLPCX without the CMV promoter and polyadenylation signal. 7SK RNA was 5′ tagged with the PP7 hairpin followed by the tobramycin aptamer (PT-7SK) or 3′ tagged with the hairpins in either possible orientation (7SK-PT, 7SK-TP). Endogenous and recombinant 7SK RNAs were detected by Northern blot hybridization using total RNA from transiently transfected 293T cells. Transfection efficiency was routinely >70% (data not shown). (C) The PT-7SK RNA expression cassette was also subcloned into the vector backbone of pRc/CMV without the CMV promoter and polyadenylation signal. Endogenous and recombinant 7SK RNAs were detected as in B.
FIGURE 2.
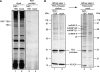
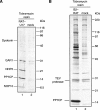
RAT purification of 7SK RNPs. (A) Purification of RAT-7SK RNA. Extracts from cells transiently transfected to express RAT-7SK or from untransfected cells were used for parallel affinity purifications. RNA was isolated from input whole cell extracts (lanes 1,2) or washed tobramycin resin (lanes 3,4), fractionated by denaturing PAGE, and visualized by staining with SYBR Gold. (B) Proteins in the TEV protease elution (lanes 1,2) or bound to washed tobramycin resin (lanes 3,4) from RAT-7SK RNA and mock purifications were resolved by SDS-PAGE and visualized by silver staining. Putative protein assignments are based on correlations to expected mobilities of proteins identified by mass spectrometry. All samples, including markers, were resolved on the same gel.
FIGURE 3.
RAT purification of U17 and B2 RNPs. (A) The human U17 small nucleolar RNA, a member of the H/ACA-motif family of snoRNAs responsible for processing of the ribosomal RNA precursor (Eliceiri 2006), was tagged at the 5′ end with hairpins in the PT order. One-tenth of the material bound to washed tobramycin resin was used for SDS-PAGE and silver staining. The remaining nine-tenths were used for protein identification by mass spectrometry, which confirmed the presence of the core H/ACA-motif binding proteins dyskerin, GAR1, NHP2, and NOP10 specifically in the RAT-U17 purification. Silver staining of dyskerin was less intense than expected in relation to the other H/ACA-motif binding proteins, but dyskerin peptide sequences were not fewer than expected in relation to the other proteins in the mass spectrometry results, suggesting that silver staining intensity was unequal across this gel. (B) The SINE-encoded mouse B2 RNA, which has been suggested to inhibit Pol II transcription in response to cellular stress (Allen et al. 2004; Espinoza et al. 2004), was tagged at the 3′ end with hairpins in the PT order.
FIGURE 4.
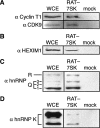
RAT purification specificity of candidate 7SK RNP proteins. Proteins in input whole cell extract (WCE) or bound to washed tobramycin resin of RAT-7SK or mock affinity purifications were used for immunoblots with the antibodies indicated.
FIGURE 5.
Protein associations with endogenous 7SK RNPs. (A) Endogenous and recombinant proteins were detected using antibodies against hnRNP Q/R (left) or hnRNP K (right). Endogenous hnRNP R, hnRNP Q isoforms, and hnRNP K isoforms are labeled. Primary antibody binding to the Protein A domains of the TAP tag amplifies the immunoblot signal of tagged proteins. However, this signal amplification can be eliminated by TEV protease cleavage between the Protein A domains and the calmodulin-binding peptide of the TAP tag. Protease-cleaved TAP-tagged hnRNP K4 is not evident due to its probable comigration with the larger isoform of endogenous hnRNP K. (B) TAP-tagged proteins in extracts of cells treated with DMSO alone (odd lanes) or DRB in DMSO (even lanes) were purified using IgG agarose. RNAs from input extracts were examined by Northern blot hybridization to detect 7SK RNA (top panel). RNAs bound to IgG agarose were examined by Northern blot hybridization to detect 7SK RNA (second panel from top) or stained with SYBR Gold following denaturing PAGE to detect endogenous RNAs (large panel) and the internal precipitation control (PC) added before RNA precipitation (bottom panel). Untransfected cell extract was used for immunopurification of cyclin T1 as a control (lanes 19,20). (C) TAP-tagged proteins in the whole cell extracts used for purification in B were detected by immunoblotting with nonspecific rabbit IgG. Asterisks mark the position of recombinant C9orf10 protein (lanes 13,14), which is equally but barely detectable in both lanes. The upper half of the blot is shown for lanes 9–16 as an overexposure relative to the bottom half to enable visualization of the C9orf10 signal.
FIGURE 6.
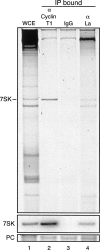
Immunopurification of cyclin T1 or La enriches 7SK RNA. Whole cell extract was incubated with immobilized antibodies against cyclin T1 or La or a nonspecific goat IgG control. RNA extracted from the input whole cell extract (lane 1) and antibody-bound material (lanes 2_–_4) was resolved by denaturing PAGE and detected by either SYBR Gold staining (top panel) or Northern blot hybridization for 7SK RNA (center panel). An internal tRNA precipitation control (PC) added during RNA extraction was visualized by SYBR Gold staining (bottom panel).
FIGURE 7.
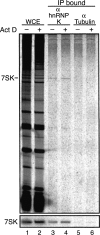
Cross-linking of hnRNP K to 7SK RNA in vivo. Untreated cells or cells treated with actinomycin D (Act D) were cross-linked with formaldehyde. Whole cell extracts (WCE) were prepared by sonication and immunopurified using antibodies against hnRNP K or tubulin. RNA in the WCE and antibody-bound fractions (IP bound) was resolved by denaturing PAGE and detected by either SYBR Gold staining (top panel) or Northern blot hybridization for 7SK RNA (bottom panel).
FIGURE 8.
Distinct, potentially multi-subunit 7SK RNPs. Whole cell extracts from untransfected cells were used for immunoaffinity purification with cyclin T1 antibody or nonspecific IgG. Whole cell extracts from cells transfected to express TAP-tagged proteins or the TAP tag alone (TAP) from the pcDNA 3.1 vector backbone were used for affinity purification with IgG agarose. (At right) Part of the input and bound fractions was used to extract RNA for SYBR Gold staining of input and precipitation controls and Northern blot hybridization to detect 7SK RNA. (A) Complexes were eluted with 2% SDS and used for immunoblotting with an antibody against CDK9. The asterisk denotes signal from nonspecific cross-reaction with TAP-hnRNP K4 due to the tag Protein A domains. All of the samples shown were run on the same gel and imaged together using the same setting. (B) Complexes were eluted with HLB150 supplemented with 1 M urea, 0.5 M NaCl, and 0.05% SDS and used for immunoblotting with antibodies against hnRNP K or hnRNP Q/R. The asterisk denotes signal from signal from nonspecific cross-reaction with TAP-hnRNP K4 due to the tag Protein A domains. The copurification of hnRNP R is obscured by TAP-hnRNP K.
FIGURE 9.
Effect of hnRNP K depletion on 7SK RNP assembly. (A) Whole cell extracts from cells transfected with hnRNP K or control siRNAs were used for immunoaffinity purification with a cyclin T1 antibody. RNAs from input and bound fractions were examined by Northern blot hybridization to detect the indicated RNAs and an oligonucleotide precipitation control (PC). The bead recovery control (BRC) was visualized by SYBR Gold staining. All samples were electrophoresed on the same gel, and identical settings were used to visualize WCE and bound signals for each RNA. (B) Quantification of data in A. 7SK RNA recovery was normalized to the precipitation control and input 7SK RNA levels. Error bars indicate the standard error of the mean from three independent samples. (C) Immunoblots of hnRNP K, cyclin T1, and HEXIM1 in the whole cell extracts used for purification in A. Note that control siRNA lanes have slightly less total protein than hnRNP K siRNA lanes. (D) Model for 7SK RNP compositions and their dynamics of exchange. P-TEFb and HEXIM proteins are illustrated in accord with previous studies (Li et al. 2005; Egloff et al. 2006). The relative placement of newly identified 7SK RNP proteins on a shared molecule of 7SK RNA is intended as an aid for visualization, not as a specific model of RNP structure.
Similar articles
- Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes.
Fouraux MA, Bouvet P, Verkaart S, van Venrooij WJ, Pruijn GJ. Fouraux MA, et al. J Mol Biol. 2002 Jul 12;320(3):475-88. doi: 10.1016/s0022-2836(02)00518-1. J Mol Biol. 2002. PMID: 12096904 - Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function.
Wassarman DA, Steitz JA. Wassarman DA, et al. Mol Cell Biol. 1991 Jul;11(7):3432-45. doi: 10.1128/mcb.11.7.3432-3445.1991. Mol Cell Biol. 1991. PMID: 1646389 Free PMC article. - Stabilize and connect: the role of LARP7 in nuclear non-coding RNA metabolism.
Hasler D, Meister G, Fischer U. Hasler D, et al. RNA Biol. 2021 Feb;18(2):290-303. doi: 10.1080/15476286.2020.1767952. Epub 2020 Jun 3. RNA Biol. 2021. PMID: 32401147 Free PMC article. Review. - Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs.
Matera AG, Terns RM, Terns MP. Matera AG, et al. Nat Rev Mol Cell Biol. 2007 Mar;8(3):209-20. doi: 10.1038/nrm2124. Nat Rev Mol Cell Biol. 2007. PMID: 17318225 Review. - Ribonucleoprotein particles containing non-coding Y RNAs, Ro60, La and nucleolin are not required for Y RNA function in DNA replication.
Langley AR, Chambers H, Christov CP, Krude T. Langley AR, et al. PLoS One. 2010 Oct 27;5(10):e13673. doi: 10.1371/journal.pone.0013673. PLoS One. 2010. PMID: 21060685 Free PMC article.
Cited by
- Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat.
Muniz L, Egloff S, Ughy B, Jády BE, Kiss T. Muniz L, et al. PLoS Pathog. 2010 Oct 14;6(10):e1001152. doi: 10.1371/journal.ppat.1001152. PLoS Pathog. 2010. PMID: 20976203 Free PMC article. - An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins.
Leppek K, Stoecklin G. Leppek K, et al. Nucleic Acids Res. 2014 Jan;42(2):e13. doi: 10.1093/nar/gkt956. Epub 2013 Oct 23. Nucleic Acids Res. 2014. PMID: 24157833 Free PMC article. - Structural basis for the coevolution of a viral RNA-protein complex.
Chao JA, Patskovsky Y, Almo SC, Singer RH. Chao JA, et al. Nat Struct Mol Biol. 2008 Jan;15(1):103-5. doi: 10.1038/nsmb1327. Epub 2007 Dec 9. Nat Struct Mol Biol. 2008. PMID: 18066080 Free PMC article. - A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination.
Koldobskaya Y, Duguid EM, Shechner DM, Suslov NB, Ye J, Sidhu SS, Bartel DP, Koide S, Kossiakoff AA, Piccirilli JA. Koldobskaya Y, et al. Nat Struct Mol Biol. 2011 Jan;18(1):100-6. doi: 10.1038/nsmb.1945. Epub 2010 Dec 12. Nat Struct Mol Biol. 2011. PMID: 21151117 Free PMC article. - An RNA matchmaker protein regulates the activity of the long noncoding RNA HOTAIR.
Meredith EK, Balas MM, Sindy K, Haislop K, Johnson AM. Meredith EK, et al. RNA. 2016 Jul;22(7):995-1010. doi: 10.1261/rna.055830.115. Epub 2016 May 4. RNA. 2016. PMID: 27146324 Free PMC article.
References
- Allen, T.A., Von Kaenel, S., Goodrich, J.A., Kugel, J.F. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004;11:816–821. - PubMed
- Ausubel, F., Brent, R., Kingston, R., Moore, D., Seidman, F., Smith, J., Struhl, K. Current protocols in molecular biology. Wiley; New York: 1996.
- Bertone, P., Gerstein, M., Snyder, M. Applications of DNA tiling arrays to experimental genome annotation and regulatory pathway discovery. Chromosome Res. 2005;13:259–274. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials