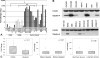Claudin-4 overexpression in epithelial ovarian cancer is associated with hypomethylation and is a potential target for modulation of tight junction barrier function using a C-terminal fragment of Clostridium perfringens enterotoxin - PubMed (original) (raw)
Comparative Study
doi: 10.1593/neo.07118.
Joseph Kwong, Chun-Min Lo, James G Smedley 3rd, Bruce A McClane, Margarita Aponte, Zhijian Gao, Jennifer L Sarno, Jennifer Hinners, William R Welch, Ross S Berkowitz, Samuel C Mok, Elizabeth I O Garner
Affiliations
- PMID: 17460774
- PMCID: PMC1854850
- DOI: 10.1593/neo.07118
Comparative Study
Claudin-4 overexpression in epithelial ovarian cancer is associated with hypomethylation and is a potential target for modulation of tight junction barrier function using a C-terminal fragment of Clostridium perfringens enterotoxin
Babak Litkouhi et al. Neoplasia. 2007 Apr.
Abstract
Background: Claudin-4, a tight junction (TJ) protein and receptor for the C-terminal fragment of Clostridium perfringens enterotoxin (C-CPE), is overexpressed in epithelial ovarian cancer (EOC). Previous research suggests DNA methylation is a mechanism for claudin-4 overexpression in cancer and that C-CPE acts as an absorption-enhancing agent in claudin-4-expressing cells. We sought to correlate claudin-4 overexpression in EOC with clinical outcomes and TJ barrier function, investigate DNA methylation as a mechanism for overexpression, and evaluate the effect of C-CPE on the TJ.
Methods: Claudin-4 expression in EOC was quantified and correlated with clinical outcomes. Claudin-4 methylation status was determined, and claudin-4-negative cell lines were treated with a demethylating agent. Electric cell-substrate impedance sensing was used to calculate junctional (paracellular) resistance (Rb) in EOC cells after claudin-4 silencing and after C-CPE treatment.
Results: Claudin-4 overexpression in EOC does not correlate with survival or other clinical endpoints and is associated with hypomethylation. Claudin-4 overexpression correlates with Rb and C-CPE treatment of EOC cells significantly decreased Rb in a dose- and claudin-4-dependent noncytotoxic manner.
Conclusions: C-CPE treatment of EOC cells leads to altered TJ function. Further research is needed to determine the potential clinical applications of C-CPE in EOC drug delivery strategies.
Figures
Figure 1
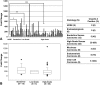
(A) qRT-PCR was used to compare claudin-4 expression in 79 serous ovarian cancer tissue samples of various grades to HOSE tissue. Claudin-4 expression was significantly higher in borderline, low-grade, and high-grade cancers when compared to HOSE. *P = .003, **P = .02, ***P < .001; (B) expression was not significantly different between high-grade and borderline or low-grade cancers (P = .14). Table: Using IHC, 76 ovarian cancer tissue samples of various histologic types were stained for claudin-4 and compared to nine HOSE and six endosalpingiosis samples. Slides were scored as follows: 0, absent/trace staining; 1, minimal staining; 2, moderate staining; 3, strong staining. Samples were categorized into “claudin-4-negative” (absent/trace staining, score 0) and “claudin-4-positive” (any degree of staining, scores 1–3) groups for comparison and statistical analysis.
Figure 2
Representative claudin-4-immunostained IHC slides from various histologic types of ovarian tissue. (A) Normal ovarian surface epithelium, (B) endosalpingiosis, (C and D) borderline serous carcinoma, (E and F) high-grade serous carcinoma, (G) clear cell carcinoma, (H) endometrioid carcinoma, and (I) mucinous carcinoma. Scale bar = 20 µm.
Figure 3
(A) qRT-PCR was used to compare claudin-4 expression in 20 ovarian cancer cell lines to eight HOSE and six immortalized HOSE cell lines. *P < .001, **P = .04. (B) Claudin-4 expression, as determined by Western blot, in ovarian cancer and HOSE cell lines. β-Actin was used as a loading control. (C) Claudin-4 expression, as determined by qRT-PCR, was compared in a subset of high-grade advanced-stage tissue samples. There was no statistically significant difference between paired primary and recurrent tumors, platinum-sensitive and platinum-resistant tumors, or short-term (less than 24 months) and long-term (60 months or more) survivors.
Figure 4
MSPCR was performed and average of three to four times on genomic DNA obtained from (A) nine claudin-4-expressing ovarian cancer tissue samples (393, 358, 377, 317, 351, 345, 312, 380, 412), (B) 11 ovarian cancer cell lines (OVCAR3, OVCA429, OVCA433, DOV13, PEO4, ES2, CAOV3, TOV112D, MCAS, RMUG-L, OVCA432) and three HOSE cell lines (HOSE2166, HOSE2170, HOSE2177). Also shown in (B), the methylation status of CLDN4 was compared to its protein expression in the cell lines. β-Actin was used as a loading control. Me, methylated primer; U-Me, unmethylated primer; WB, Western blot.
Figure 5
Ovarian cancer cell lines DOV13 and TOV112D, neither of which expressed claudin-4, were treated with 5-AZA (1 µmol/l), an inhibitor of DNA methylation. Treatment with 5-AZA resulted in the appearance of new unmethylated MSPCR products (inset) and corresponded with increased expression of claudin-4, as assessed by qRT-PCR (bar graph). The experiments were performed in duplicate. Also shown, MSPCR resulted in a methylated product for the methylated control DNA; PCR products were absent for unmodified DNA (which had not undergone bisulfite treatment). Me, methylated primer; U-Me, unmethylated primer.
Figure 6
(A) The schematic diagram represents claudin-4 mRNA: +1, transcription initiation site; | (vertical marks), CpG dinucleotide; blue band, CpG island identified using CpG Island Searcher software (
http://ccnt.hsc.usc.edu/cpgislands/
) and the displayed criteria; → (arrowhead), forward (BSF, bp 156–180) and reverse (BSR, bp 468–491) primer locations for the bisulfite sequencing; ▸ (arrows), forward (MSPF, bp 233–256) and reverse (MSPR, bp 432–452) primer locations for MSPCR. (B) The ovarian cancer cell line OVCA429, which exhibited a hypomethylated CLDN4 allele only (per Figure 4_B_), underwent bisulfite sequencing. Complete hypomethylation was demonstrated at all 21 CpG dinucleotides (circled and numbered) in the sequenced fragment.
Figure 7
(A) Claudin-4 Western blot for SKOV3 WT, a negative control, and SKOV3 siRNA (claudin-4 silenced). β-Actin was used as a loading control. (B) Junctional (paracellular) resistance (Rb) was compared between SKOV3 WT (wild type) and SKOV3 siRNA (siRNA claudin-4 silenced) cell lines over 7 days in cell culture medium, *P < .05. Cell lines (C) OVCA429 and (D) SKOV3 were treated with increasing amounts of C-CPE (µg/µl), resulting in a dose-dependent decrease in Rb compared to control (cell culture medium only, C-CPE concentration of 0 µg/µl). Bars represent means ± SD. Measurements were repeated four to six times, *P < .05.
Similar articles
- C terminus of Clostridium perfringens enterotoxin downregulates CLDN4 and sensitizes ovarian cancer cells to Taxol and Carboplatin.
Gao Z, Xu X, McClane B, Zeng Q, Litkouhi B, Welch WR, Berkowitz RS, Mok SC, Garner EI. Gao Z, et al. Clin Cancer Res. 2011 Mar 1;17(5):1065-74. doi: 10.1158/1078-0432.CCR-10-1644. Epub 2010 Dec 1. Clin Cancer Res. 2011. PMID: 21123456 Free PMC article. - Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: Evidence for direct involvement of claudins in tight junction barrier.
Sonoda N, Furuse M, Sasaki H, Yonemura S, Katahira J, Horiguchi Y, Tsukita S. Sonoda N, et al. J Cell Biol. 1999 Oct 4;147(1):195-204. doi: 10.1083/jcb.147.1.195. J Cell Biol. 1999. PMID: 10508866 Free PMC article. - Cytotoxicity of Clostridium perfringens enterotoxin depends on the conditions of claudin-4 in ovarian carcinoma cells.
Tanaka S, Aoyama T, Ogawa M, Takasawa A, Murata M, Osanai M, Saito T, Sawada N. Tanaka S, et al. Exp Cell Res. 2018 Oct 1;371(1):278-286. doi: 10.1016/j.yexcr.2018.08.024. Epub 2018 Aug 22. Exp Cell Res. 2018. PMID: 30142326 - A novel strategy for a drug delivery system using a claudin modulator.
Kondoh M, Takahashi A, Fujii M, Yagi K, Watanabe Y. Kondoh M, et al. Biol Pharm Bull. 2006 Sep;29(9):1783-9. doi: 10.1248/bpb.29.1783. Biol Pharm Bull. 2006. PMID: 16946486 Review. - [Non-invasive drug delivery system with the claudin binder].
Takahashi A, Kondoh M, Yagi K. Takahashi A, et al. Yakugaku Zasshi. 2011;131(11):1583-7. doi: 10.1248/yakushi.131.1583. Yakugaku Zasshi. 2011. PMID: 22041696 Review. Japanese.
Cited by
- Clostridium perfringens enterotoxin (CPE) and CPE-binding domain (c-CPE) for the detection and treatment of gynecologic cancers.
Black JD, Lopez S, Cocco E, Schwab CL, English DP, Santin AD. Black JD, et al. Toxins (Basel). 2015 Apr 1;7(4):1116-25. doi: 10.3390/toxins7041116. Toxins (Basel). 2015. PMID: 25835384 Free PMC article. Review. - Epigenomics and ovarian carcinoma.
Maldonado L, Hoque MO. Maldonado L, et al. Biomark Med. 2010 Aug;4(4):543-70. doi: 10.2217/bmm.10.72. Biomark Med. 2010. PMID: 20701443 Free PMC article. Review. - Specificity of interaction between clostridium perfringens enterotoxin and claudin-family tight junction proteins.
Mitchell LA, Koval M. Mitchell LA, et al. Toxins (Basel). 2010 Jul;2(7):1595-611. doi: 10.3390/toxins2071595. Epub 2010 Jun 24. Toxins (Basel). 2010. PMID: 22069652 Free PMC article. Review. - Genome-wide DNA methylation identifies trophoblast invasion-related genes: Claudin-4 and Fucosyltransferase IV control mobility via altering matrix metalloproteinase activity.
Hu Y, Blair JD, Yuen RK, Robinson WP, von Dadelszen P. Hu Y, et al. Mol Hum Reprod. 2015 May;21(5):452-65. doi: 10.1093/molehr/gav007. Epub 2015 Feb 19. Mol Hum Reprod. 2015. PMID: 25697377 Free PMC article. - Validity of Xiphophorus fish as models for human disease.
Schartl M, Lu Y. Schartl M, et al. Dis Model Mech. 2024 Jan 1;17(1):dmm050382. doi: 10.1242/dmm.050382. Epub 2024 Feb 1. Dis Model Mech. 2024. PMID: 38299666 Free PMC article. Review.
References
- Bonome T, Lee JY, Park DC, Radonovich M, Pise-Masison C, Brady J, Gardner GJ, Hao K, Wong WH, Barrett JC, et al. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res. 2005;65:10602–10612. - PubMed
- Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, Cho KR, Riggins GJ, Morin PJ. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000;60:6281–6287. - PubMed
- Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, Au JL, Rosen DG, Liu J, Hellstrom I, et al. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10:3291–3300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R33 CA103595/CA/NCI NIH HHS/United States
- R37 AI019844/AI/NIAID NIH HHS/United States
- P50 CA105009/CA/NCI NIH HHS/United States
- R37AI19844/AI/NIAID NIH HHS/United States
- R33CA103595/CA/NCI NIH HHS/United States
- P50CA105009/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous