Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes - PubMed (original) (raw)
Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes
Yi Huang et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Epigenetic chromatin modification is a major regulator of eukaryotic gene expression, and aberrant epigenetic silencing of gene expression contributes to tumorigenesis. Histone modifications include acetylation, phosphorylation, and methylation, resulting in a combination of histone marks known collectively as the histone code. The chromatin marks at a given promoter determine, in part, whether specific promoters are in an open/active conformation or closed/repressed conformation. Dimethyl-lysine 4 histone H3 (H3K4me2) is a transcription-activating chromatin mark at gene promoters, and demethylation of this mark by the lysine-specific demethylase 1 (LSD1), a homologue of polyamine oxidases, may broadly repress gene expression. We now report that novel biguanide and bisguanidine polyamine analogues are potent inhibitors of LSD1. These analogues inhibit LSD1 in human colon carcinoma cells and affect a reexpression of multiple, aberrantly silenced genes important in the development of colon cancer, including members of the secreted frizzle-related proteins (SFRPs) and the GATA family of transcription factors. Furthermore, we demonstrate by chromatin immunoprecipitation analysis that the reexpression is concurrent with increased H3K4me2 and acetyl-H3K9 marks, decreased H3K9me1 and H3K9me2 repressive marks. We thus define important new agents for reversing aberrant repression of gene transcription.
Conflict of interest statement
Conflict of interest statement: With respect to competing financial interests, a patent application has been filed by P.M.W. and R.A.C. to cover the compounds reported in this manuscript. As of January 2007, P.M.W. and R.A.C. serve as consultants to Cellgate, Inc., which has an option to license these compounds.
Figures
Fig. 1.
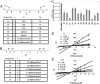
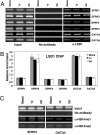
Inhibition of LSD1 by polyamine analogues. (A) Bisguanidines (1a–1g). (B) Biguanides (2a–2f). (C) Three micrograms of purified LSD1 protein were incubated with 5 μM H3K4me2 (1–21 aa) as substrate in the presence of 1 μM of the indicated analogue. The results represent the mean of three determinations ± SD. (D and E) The effects of increasing concentrations of 1c (D) or 2d (E) on LSD1 activity in the presence of increasing substrate concentrations. Double reciprocal plots indicate inhibition of LSD1 by 1c and 2d to be noncompetitive.
Fig. 2.
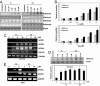
Inhibition of LSD1 by polyamine analogues increases global H3K4me1 and H3K4me2, resulting in reexpression of aberrantly silenced genes in treated human colon cancer cells. (A) HCT116 cells were exposed to increasing concentrations of the indicated compound for 48 h, and 30 μg of nuclear protein per lane were analyzed for expression of H3K4me1, H3K4me2, H3K9me2, and PCNA as a loading control. (B) Histograms represent the mean protein expression levels of three determinations relative to PCNA ± SD, as determined by quantitative immunoblotting using infrared detection and analysis. (C) HCT116 cells were treated for 48 h with the indicated compounds, and total RNA was extracted for RT-PCR analysis of SFRP1, SFRP4, SFRP5, and GATA5 expression. GAPDH is included as an internal control. The results shown are from a single experiment repeated at least three times with similar results. (D) RKO cells were exposed to increasing concentrations of the indicated compound for 48 h, and 30 μg of nuclear protein per lane were analyzed for expression of H3K4me2 and PCNA. The histogram represents the mean protein amount of three determinations relative to PCNA ± SD. (E) RKO cells were treated for 48 h with the indicated compounds and total RNA was extracted for RT-PCR analysis of SFRP4 and SFRP5 expression. GAPDH is included as an internal control. The results shown are from a single experiment repeated at least three times with similar results.
Fig. 3.
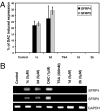
Relative reexpression of SFRP4 and SFRP5 induced by polyamine analogue inhibitors of LSD1. HCT116 cells were treated with 5 μM 1c, 2d, 1d, or 2b; 1 μM DAC; or 300 nM TSA for 48 h. (A) qPCR analysis of SFRP4 and SFRP5 expression. Results are presented relative to expression induced by DAC and represent the mean of three independent experiments, each performed in triplicate ± SD. (B) Representative qPCR products were analyzed on GelStar-stained 2% agarose gel.
Fig. 4.
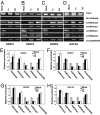
Inhibition of LSD1 by polyamine analogues increases activating H3K4me2 and acetyl H3K9 marks and decreases repressive H3K9me1 and H3K9me2 marks at the promoters of reexpressed genes. HCT116 cells were treated with 5 μM of the indicated compound for 48 h. (A–D) ChIP analysis was used to determine the occupancy of the indicated promoters by multiple activating and repressive marks. (E–H) The quantified results are the means of three independent experiments with an SD as indicated.
Fig. 5.
Promoter occupancy by LSD1. (A) HCT116 cells were treated with 5 μM of the indicated compounds for 48 h. ChIP analysis was performed to determine the occupancy of the indicated promoters by LSD1. (B) The quantified results are the mean of three independent experiments ± SD, relative to LSD1 detected at the SFRP1 promoter of untreated cells. (C) After HCT116 cells were treated with 5 μM of the indicated compounds for 48 h, ChIP was used to determine the levels of H3K4me2 and H3K4me1 at the promoters of SFRP2 and GATA4.
Fig. 6.
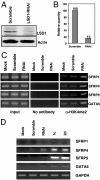
Knockdown of LSD1 by siRNA leads to specific gene reexpression. (A) HCT116 cells were transfected with scrambled or LSD1-targeted siRNA oligonucleotides for 48 h. Proteins isolated from transfected cells were subjected to quantitative immunoblotting with an antibody to LSD1. (B) The histogram represents the relative LSD1 protein quantity in scrambled and LSD1 siRNA transfectants. (C) ChIP analysis was used to determine the levels of H3K4me2 in the promoters of indicated genes. (D) HCT116 cells were treated for 48 h with 5 μM 1c or 2d or transfected with scrambled or LSD1 siRNA oligonucleotides for 48 h. RNA was extracted for RT-PCR analysis of expression of the indicated genes. GAPDH is included as an internal control.
Similar articles
- Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes.
Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, Jones RJ, Woster PM, Casero RA Jr. Huang Y, et al. Clin Cancer Res. 2009 Dec 1;15(23):7217-28. doi: 10.1158/1078-0432.CCR-09-1293. Epub 2009 Nov 24. Clin Cancer Res. 2009. PMID: 19934284 Free PMC article. - The re-expression of the epigenetically silenced e-cadherin gene by a polyamine analogue lysine-specific demethylase-1 (LSD1) inhibitor in human acute myeloid leukemia cell lines.
Murray-Stewart T, Woster PM, Casero RA Jr. Murray-Stewart T, et al. Amino Acids. 2014 Mar;46(3):585-94. doi: 10.1007/s00726-013-1485-1. Epub 2013 Mar 19. Amino Acids. 2014. PMID: 23508577 Free PMC article. - Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state.
McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. McGarvey KM, et al. Cancer Res. 2006 Apr 1;66(7):3541-9. doi: 10.1158/0008-5472.CAN-05-2481. Cancer Res. 2006. PMID: 16585178 - Polyamine analogues targeting epigenetic gene regulation.
Huang Y, Marton LJ, Woster PM, Casero RA. Huang Y, et al. Essays Biochem. 2009 Nov 4;46:95-110. doi: 10.1042/bse0460007. Essays Biochem. 2009. PMID: 20095972 Free PMC article. Review. - New roles of flavoproteins in molecular cell biology: histone demethylase LSD1 and chromatin.
Forneris F, Battaglioli E, Mattevi A, Binda C. Forneris F, et al. FEBS J. 2009 Aug;276(16):4304-12. doi: 10.1111/j.1742-4658.2009.07142.x. Epub 2009 Jul 14. FEBS J. 2009. PMID: 19624733 Review.
Cited by
- Epigenetic Reexpression of Hemoglobin F Using Reversible LSD1 Inhibitors: Potential Therapies for Sickle Cell Disease.
Holshouser S, Cafiero R, Robinson M, Kirkpatrick J, Casero RA Jr, Hyacinth HI, Woster PM. Holshouser S, et al. ACS Omega. 2020 Jun 9;5(24):14750-14758. doi: 10.1021/acsomega.0c01585. eCollection 2020 Jun 23. ACS Omega. 2020. PMID: 32596612 Free PMC article. - Inhibition of FAD-dependent lysine-specific demethylases by chiral polyamine analogues.
Umezawa N, Tsuji K, Sato S, Kikuchi M, Watanabe H, Horai Y, Yamaguchi M, Hisamatsu Y, Umehara T, Higuchi T. Umezawa N, et al. RSC Adv. 2018 Oct 31;8(64):36895-36902. doi: 10.1039/c8ra07879c. eCollection 2018 Oct 26. RSC Adv. 2018. PMID: 35558920 Free PMC article. - Identification of four potential epigenetic modulators from the NCI structural diversity library using a cell-based assay.
Best AM, Chang J, Dull AB, Beutler JA, Martinez ED. Best AM, et al. J Biomed Biotechnol. 2011;2011:868095. doi: 10.1155/2011/868095. Epub 2010 Dec 22. J Biomed Biotechnol. 2011. PMID: 21234371 Free PMC article. - Epigenetics: Tools and Technologies.
Janzen WP, Wigle TJ, Jin J, Frye SV. Janzen WP, et al. Drug Discov Today Technol. 2010 Spring;7(1):e59-e65. doi: 10.1016/j.ddtec.2010.07.004. Drug Discov Today Technol. 2010. PMID: 21243036 Free PMC article. - Mechanisms of carbon nanotube aggregation and the reversion of carbon nanotube aggregates in aqueous medium.
Koh B, Cheng W. Koh B, et al. Langmuir. 2014 Sep 16;30(36):10899-909. doi: 10.1021/la5014279. Epub 2014 Sep 4. Langmuir. 2014. PMID: 25144606 Free PMC article.
References
- Jenuwein T, Allis CD. Science. 2001;293:1074–1080. - PubMed
- Jaenisch R, Bird A. Nat Genet. 2003;33(Suppl):245–254. - PubMed
- Baylin SB, Ohm JE. Nat Rev Cancer. 2006;6:107–116. - PubMed
- Lachner M, O'Sullivan RJ, Jenuwein T. J Cell Sci. 2003;116:2117–2124. - PubMed
- Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Nat Rev Cancer. 2001;1:194–202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA43318/CA/NCI NIH HHS/United States
- R01 CA051085/CA/NCI NIH HHS/United States
- ES11858/ES/NIEHS NIH HHS/United States
- R01 CA098454/CA/NCI NIH HHS/United States
- R01 ES011858/ES/NIEHS NIH HHS/United States
- CA51085/CA/NCI NIH HHS/United States
- CA98454/CA/NCI NIH HHS/United States
- R01 CA043318/CA/NCI NIH HHS/United States
- R01 CA085509/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources