Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes - PubMed (original) (raw)
. 2007 Jun 1;316(5829):1336-41.
doi: 10.1126/science.1142364. Epub 2007 Apr 26.
Michael N Weedon, Cecilia M Lindgren, Timothy M Frayling, Katherine S Elliott, Hana Lango, Nicholas J Timpson, John R B Perry, Nigel W Rayner, Rachel M Freathy, Jeffrey C Barrett, Beverley Shields, Andrew P Morris, Sian Ellard, Christopher J Groves, Lorna W Harries, Jonathan L Marchini, Katharine R Owen, Beatrice Knight, Lon R Cardon, Mark Walker, Graham A Hitman, Andrew D Morris, Alex S F Doney; Wellcome Trust Case Control Consortium (WTCCC); Mark I McCarthy, Andrew T Hattersley
Affiliations
- PMID: 17463249
- PMCID: PMC3772310
- DOI: 10.1126/science.1142364
Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes
Eleftheria Zeggini et al. Science. 2007.
Erratum in
- Science. 2007 Aug 24;317(5841):1035-6
Abstract
The molecular mechanisms involved in the development of type 2 diabetes are poorly understood. Starting from genome-wide genotype data for 1924 diabetic cases and 2938 population controls generated by the Wellcome Trust Case Control Consortium, we set out to detect replicated diabetes association signals through analysis of 3757 additional cases and 5346 controls and by integration of our findings with equivalent data from other international consortia. We detected diabetes susceptibility loci in and around the genes CDKAL1, CDKN2A/CDKN2B, and IGF2BP2 and confirmed the recently described associations at HHEX/IDE and SLC30A8. Our findings provide insight into the genetic architecture of type 2 diabetes, emphasizing the contribution of multiple variants of modest effect. The regions identified underscore the importance of pathways influencing pancreatic beta cell development and function in the etiology of type 2 diabetes.
Figures
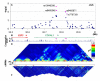
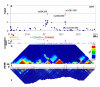
Figure 1. Overview of CDKAL1 signal region
A Plot of −log(p values) for T2D (Cochran-Armitage test for trend) against chromosome position in Mb. Blue diamonds represent primary scan results and pink triangles denote meta-analysis results across all UK samples. B Genomic location of genes showing intron and exon structure (NCBI Build 35). Pink triangles show position of replication SNPs relative to gene structure. C MULTIZ (24) vertebrate alignment of 17 species showing evolutionary conservation. D GoldSurfer2 (25) plot of linkage disequilibrium (r2) for SNPs genotyped in WTCCC scan (passing T2D-specific quality control) in WTCCC T2D cases. E Recombination rate given as cM/MB. Red boxes represent recombination hotspots (26). F GoldSurfer2 plot of linkage disequilibrium (r2) for all HapMap SNPs across the region (HapMap CEU data).
Figure 2. Overview of chr9 signal region
Panel layout as per Figure 1
References
- Stumvoll M, Goldstein BJ, van Haeften TW. Lancet. 2005;365:1333–1346. - PubMed
- Altshuler D, et al. Nat. Genet. 2000;26:76–80. - PubMed
- Gloyn AL, et al. Diabetes. 2003;52:568–572. - PubMed
- Grant SF, et al. Nat. Genet. 2006;38:320–323. - PubMed
- Donnelly P, the WTCCC, personal communication Data from the Wellcome Trust Case Control Consortium scan.
Publication types
MeSH terms
Substances
Grants and funding
- 083948/WT_/Wellcome Trust/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- G0000934/MRC_/Medical Research Council/United Kingdom
- G0500070/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous