Inhibition of the ubiquitin-proteasome system induces stress granule formation - PubMed (original) (raw)
Inhibition of the ubiquitin-proteasome system induces stress granule formation
Rachid Mazroui et al. Mol Biol Cell. 2007 Jul.
Abstract
The inhibition of the ubiquitin-dependent proteasome system (UPS) via specific drugs is one type of approach used to combat cancer. Although it has been suggested that UPS inhibition prevents the rapid decay of AU-rich element (ARE)-containing messages, very little is known about the cellular mechanisms leading to this effect. Here we establish a link between the inhibition of UPS activity, the formation of cytoplasmic stress granules (SGs), and mRNA metabolism. The assembly of the SGs requires the phosphorylation of the translation initiation factor eIF2alpha by a mechanism involving the stress kinase GCN2. On prolonged UPS inhibition and despite the maintenance of eIF2alpha phosphorylation, SGs disassemble and translation recovers in an Hsp72 protein-dependent manner. The formation of these SGs coincides with the disassembly of processing bodies (PBs), known as mRNA decay entities. As soon as the SGs assemble, they recruit ARE-containing messages such as p21(cip1) mRNA, which are stabilized under these conditions. Hence, our findings suggest that SGs could be considered as one of the players that mediate the early response of the cell to proteasome inhibitors by interfering temporarily with mRNA decay pathways.
Figures
Figure 1.
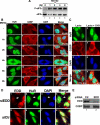
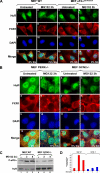
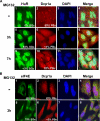
Inhibition of the proteasome activity induces SGs formation. (A) Treatment of HeLa cells with MG132 induces eIF2α phosphorylation. HeLa cells were treated with 10 μM MG132 for the indicated amounts of time, and protein extracts were prepared and analyzed by Western blotting using an anti-phospho eIF2α (top panel) or pan anti-eIF2α (bottom panel) antibodies. (B) The SGs induced by MG132 disassemble 6 h after treatment. Untreated HeLa cells or cells treated with 10 μM MG132 for different time points were permeabilized and fixed. The presence of stress granules was subsequently verified by immunofluorescence using antibodies against two surrogate markers of SGs: HuR and G3BP. The primary antibodies used were a monoclonal anti-HuR antibody and a polyclonal anti-G3BP antibody. (C) Lactacystin-induced SGs are dynamic. HeLa cells were incubated with lactacystin (10 μM) for 4 h (Lacta) or for 3 h before adding cycloheximide (50 μg/ml) for an additional hour (Lacta + CHX), and then the distribution of HuR and G3BP was analyzed as described above. In B and C the percentage of cells harboring SGs (>5 granules/cell), from three different fields and three different experiments containing a total of 450 cells, is indicated to the right bottom of 4, 8, 12, and 16. (D and E) The RNAi-mediated depletion of the E3 ubiquitin ligase EDD induces SG formation. HeLa cells transfected with siRNA against EDD or control siRNA (Ctr) for 48 h were either fixed on coverslips and used for immunofluorescence with anti-EDD and anti-HuR antibodies (D) or used to prepare total cell extracts to perform Western blot analysis using the indicated antibodies (E). (D) Cells containing SGs are indicated with arrowheads. Representative images of two independent experiments are shown.
Figure 2.
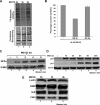
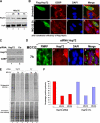
The proteasome inhibitor MG132 induces a transient reduction in translation activity that correlates with SG assembly. (A and B) MG132 triggers a transient translation inhibition. (A) HeLa cells were treated with 10 μM MG132 for the indicated times and were then labeled for 30 min with [35S]methionine (50 μCi/ml). Proteins were resolved by SDS-polyacrylamide gel, stained with Coomassie blue (bottom panel), and detected by autoradiography (top panel). (B) The intensity of the signal in each lane was measured using ImageQuant software. The results were plotted as bar graphs. Error bars, SD of three independent experiments. The percentage of each signal was defined as the intensity of each sample minus the background level and normalized against the signal for the untreated cells. (C and D) Treatment with MG132 for more than 8 h is still effective. (C) GFPu cells (293 cells stably transfected with a GFP reporter construct fused to a proteasome targeting sequence) were treated with 10 μM MG132 for different periods of time. The expression of GFP protein was monitored by Western blot using the anti-GFP antibody to assess the inhibition of the proteasome activity. (D) HeLa cells were treated with 10 μM MG132 for different periods of time (0–24 h). The expression of p53 protein was monitored by Western blot using the anti-p53 antibody. (E) The expression levels of β-actin, G3BP, HuR, and FXR1 proteins are not affected during MG132 treatment. HeLa cells were treated with MG132 as in A. The steady state level of the indicated proteins was analyzed by Western blot using specific antibodies as indicated.
Figure 3.
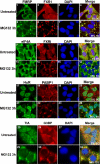
MG132-induced SGs recruit a variety of RNA-binding proteins. Untreated HeLa cells or cells treated with 10 μM MG132 for 3 h were subjected to immunofluorescence using antibodies specific to FMRP, FXR1, eIF4A, HuR, PABP, TIA, and G3BP. The Merge represents a staining for the two indicated proteins and the DAPI. The percentage of cells harboring SGs (>5 granules/cell), from three different fields and three different experiments containing a total of 450 cells, is indicated to the right bottom of 4, 8, 12, and 16. Representative pictures are shown for each protein.
Figure 4.
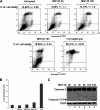
A small percentage of apoptotic cells are detected only after prolonged exposure (>15 h) to 10 μM MG132. (A and B) The effect of MG132 treatment on the expression of annexin V. (A) Untreated HeLa cells or cells that were treated with MG132 for the indicated times were collected, stained with annexin V-FITC and PI, and analyzed by flow cytometry. Formaldehyde treatment for 30 min was used as a positive control for apoptosis. The percentage of total dead or dying cells is indicated on the top of each box (% of cell death) and was defined as the sum of early (lower right box) and late (upper right box) apoptosis. The values were normalized to control, untreated cells. (B) Histogram presenting the results from A as the means ± SEM, from two independent experiments. (C) The cleavage of the apoptotic protein caspase-3 is detected only upon 16 h of exposure to MG132. HeLa cells were treated for the various indicated time points with 10 μM MG132 and then used to prepare total cell extracts. Thirty micrograms from each extract were used for Western blot, and the membrane was probed for caspase-3 and G3BP. The molecular weight (MW) of the caspase-3 cleavage product (Caspase-3-CP) is ∼14 kDa, whereas the precursor is 35 kDa. Shown are representatives of two independent experiments.
Figure 5.
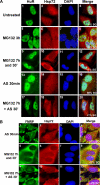
The formation of stress granules in response to the inhibition of the proteasome requires activation of the stress-induced kinase GCN2 and phosphorylation of eIF2α. (A) The phosphorylatable isoform of eIF2α is required for MG132-induced SGs. Wild-type cells as well as cells expressing a nonphosphorylatable eIF2α isoform (S51A/S51A) were treated with 10 μM MG132 for 3 h. The formation of SGs was then verified by immunofluorescence using antibodies against HuR and FXR1. (B) MG132-mediated SGs require the normal expression of the GCN2 kinase. WT MEFs or MEFs lacking GCN2 or PERK were treated with 10 μM MG132 for 3 h. The formation of SGs was verified by immunofluorescence as indicated above. (C and D) MG132-mediated eIF2α phosphorylation requires the GCN2 kinase. (C) Western blot analysis of phospho-eIF2α in wild-type as well as in GCN2−/− cells treated with 10 μM MG132 for the indicated periods of time. HuR was used as a loading control. (D) The results of Western blot were quantified and normalized against HuR. The bar graph represents the results of two independent experiments.
Figure 6.
Link between the cellular localization of the Hsp72 protein, the disassembly of SGs and translation reactivation during prolonged UPS inhibition. (A) The expression level of Hsp72 protein is enhanced during UPS inhibition. HeLa cells were treated with or without 10 μM MG132 for the indicated periods of time. The expression of Hsp72 was then analyzed by Western blot. (B) The overexpression of Hsp72 prevents SG formation. HeLa cells transfected with a construct encoding Flag-Hsp72 were treated with or without 10 μM MG132 for 3 h as indicated. The localization of Flag-Hsp72 was verified by immunofluorescence using the anti-Flag antibody. The formation of SGs was verified by immunofluorescence using antibodies against G3BP. Shown are representative images of three independent experiments. Cells overexpressing Flag-Hsp72 and did not form SGs are indicated with arrowheads. (C and D) RNAi-mediated Hsp72 depletion prevents SG disassembly after 7 h of treatment with MG132. HeLa cells incubated with an siRNA targeting Hsp72 or a control siRNA (Ctr) for 48 h were subsequently treated with 10 μM MG132 for 7 h and then lysed, and protein extracts were prepared and analyzed by Western blot for Hsp72 expression (C, HuR levels are included as a loading control). (D) The same experiment as in C was performed, and cells were fixed and used for immunofluorescence experiment with anti-FMRP and anti-Hsp72 antibodies. Cells that did not uptake the Hsp72-siRNA and lacking SGs are indicated with arrowheads. Representative images of two independent experiments are shown. (E and F) The knockdown of Hsp72 correlates with the absence of translation recovery upon treatment with MG132 for 7 h. (E) HeLa cells incubated with an siRNA targeting Hsp72, or a control siRNA, were treated with or without 10 μM MG132 for 3 or 7 h (h) and then subjected to 35S-methionine labeling for 30 min to monitor de novo protein translation. Proteins were resolved by SDS-polyacrylamide gel, stained with Coomassie blue (bottom panel), and detected by autoradiography (top panel). (F) The results were plotted as a bar graph. The intensity of the signals was defined as described in Figure 2B.
Figure 7.
Prolonged inhibition of UPS induces cytoplasmic accumulation of Hsp72 and renders the cell resistant to SG formation. (A) Inhibition of arsenite-induced HuR recruitment to SGs upon exposure to MG132 for 7 h. HeLa cells were either untreated or treated with 10 μM MG132 for 3 h, 7 h and 30 min, or 7 h with MG132 followed by 30-min exposure to 0.5 mM of arsenite (AS) and then analyzed by immunostaining with antibodies against HuR and Hsp72. HeLa cells treated with only 0.5 mM arsenite were used as a positive control for AS-induced SGs. Shown are representative images from four independent experiments. (B) Inhibition of arsenite-induced FMRP recruitment to SGs upon exposure to MG132 for 7 h. HeLa cells were treated as described in A except that SGs were visualized by following the cellular distribution of FMRP protein. Shown are representative images from three independent experiments. In both panels, the percentage of cells harboring SGs were determined as described in Figure 1.
Figure 8.
MG132-induced SGs recruit the ARE-containing message p21cip1. On treatment with MG132 for 3 h, cells were fixed, permeabilized, and then incubated with a Cy3-labeled in vitro–transcribed antisense RNA probe to detect p21cip1 mRNA (p21 mRNA probe, panels 1–8) and as a control with sense RNA probe (Ctr probe, panels 9–16). SGs were visualized using anti-HuR antibodies. Shown are representative images from two independent experiments.
Figure 9.
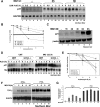
The inhibition of the proteasome blocks both ARE-mediated decay and NMD. (A and B) Effect of MG132 treatment on the half-life of p21cip1 mRNA in HeLa cells. (A) HeLa cells were first treated with or without MG132 for 3 h and then incubated with 5 μg/ml actinomycin D (ActD) for the indicated times. Total RNA was prepared and the expression levels of p21cip1 mRNA was determined using Northern blot analysis. Endogenous GAPDH mRNA was used as a loading control. (B) The expression of p21cip1 mRNA was quantified using the ImageQuant software program. The expression levels of p21cip1 mRNA were then standardized against GAPDH message and plotted as the percentage of remaining mRNA compared with message levels at the 0 time point (where there is 100% maximum mRNA level). Error bars, SD of three independent experiments. (C) Effect of MG132-treatment on the synthesis of the p21cip1 protein in HeLa cells. Total cell extracts prepared from HeLa cells treated with 10 μM MG132 for the indicated time periods were harvested and used for Western blot analysis. The membrane was probed for p21cip1 and β-actin proteins. (D and E) The GCN2 kinase seems to be required for the MG132-mediated stabilization of the p21cip1 mRNA. (D) Wild-type MEFs and MEFs derived from GCN2−/− mice were treated as in A, and the expression levels of mouse p21cip1 mRNA was determined using Northern blot analysis. (E) The expression of p21cip1 mRNA was quantified as in A. Levels were standardized against 18S rRNA. Error bars, SD of two independent experiments. (F and G) UPS inhibition by MG132 interferes with the nonsense-mediated decay pathway. (F) HeLa cells stably expressing T-cell receptor beta minigene wild type (TCR-β-wt) or a TCR-β minigene containing a PTC were treated with 10 μM MG132 for different time points, and the levels of TCR-β mRNA were analyzed by Northern blot. (G) TCR-β mRNA levels were standardized as described in A and B. Error bars, SD of two independent experiments.
Figure 10.
The inhibition of the UPS disrupts PBs. (A) After incubation with 10 μM MG132 for 3 or 7 h, HeLa cells were fixed and used for immunofluorescence to detect SGs with anti-HuR antibodies and to detect PBs with anti-Dcp1a antibodies. (B) Untreated HeLa cells or cells treated with MG132 for 3 h were analyzed by immunostaining using anti-eIF4E and anti-Dcp1a antibodies. (A and B) Shown are representative images for three independent experiments. The percentage of SGs and PBs in each cell has been determined as described in Figure 1 for SGs.
Similar articles
- Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae.
Grousl T, Ivanov P, Frýdlová I, Vasicová P, Janda F, Vojtová J, Malínská K, Malcová I, Nováková L, Janosková D, Valásek L, Hasek J. Grousl T, et al. J Cell Sci. 2009 Jun 15;122(Pt 12):2078-88. doi: 10.1242/jcs.045104. Epub 2009 May 26. J Cell Sci. 2009. PMID: 19470581 - Sorafenib, a multikinase inhibitor, induces formation of stress granules in hepatocarcinoma cells.
Adjibade P, St-Sauveur VG, Quevillon Huberdeau M, Fournier MJ, Savard A, Coudert L, Khandjian EW, Mazroui R. Adjibade P, et al. Oncotarget. 2015 Dec 22;6(41):43927-43. doi: 10.18632/oncotarget.5980. Oncotarget. 2015. PMID: 26556863 Free PMC article. - Cellular acidosis inhibits assembly, disassembly, and motility of stress granules.
Chudinova EM, Nadezhdina ES, Ivanov PA. Chudinova EM, et al. Biochemistry (Mosc). 2012 Nov;77(11):1277-84. doi: 10.1134/S0006297912110065. Biochemistry (Mosc). 2012. PMID: 23240565 - Stress granules: sites of mRNA triage that regulate mRNA stability and translatability.
Kedersha N, Anderson P. Kedersha N, et al. Biochem Soc Trans. 2002 Nov;30(Pt 6):963-9. doi: 10.1042/bst0300963. Biochem Soc Trans. 2002. PMID: 12440955 Review. - RNA granules: the good, the bad and the ugly.
Thomas MG, Loschi M, Desbats MA, Boccaccio GL. Thomas MG, et al. Cell Signal. 2011 Feb;23(2):324-34. doi: 10.1016/j.cellsig.2010.08.011. Epub 2010 Sep 8. Cell Signal. 2011. PMID: 20813183 Free PMC article. Review.
Cited by
- MDA5 localizes to stress granules, but this localization is not required for the induction of type I interferon.
Langereis MA, Feng Q, van Kuppeveld FJ. Langereis MA, et al. J Virol. 2013 Jun;87(11):6314-25. doi: 10.1128/JVI.03213-12. Epub 2013 Mar 27. J Virol. 2013. PMID: 23536668 Free PMC article. - Post-Transcriptional Regulation of Alpha One Antitrypsin by a Proteasome Inhibitor.
Rao L, Xu Y, Reineke LC, Bhattacharya A, Tyryshkin A, Shin JN, Eissa NT. Rao L, et al. Int J Mol Sci. 2020 Jun 17;21(12):4318. doi: 10.3390/ijms21124318. Int J Mol Sci. 2020. PMID: 32560429 Free PMC article. - Sam68 relocalization into stress granules in response to oxidative stress through complexing with TIA-1.
Henao-Mejia J, He JJ. Henao-Mejia J, et al. Exp Cell Res. 2009 Nov 15;315(19):3381-95. doi: 10.1016/j.yexcr.2009.07.011. Epub 2009 Jul 14. Exp Cell Res. 2009. PMID: 19615357 Free PMC article. - Stress granules, P-bodies and cancer.
Anderson P, Kedersha N, Ivanov P. Anderson P, et al. Biochim Biophys Acta. 2015 Jul;1849(7):861-70. doi: 10.1016/j.bbagrm.2014.11.009. Epub 2014 Dec 5. Biochim Biophys Acta. 2015. PMID: 25482014 Free PMC article. Review. - Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules.
Bounedjah O, Desforges B, Wu TD, Pioche-Durieu C, Marco S, Hamon L, Curmi PA, Guerquin-Kern JL, Piétrement O, Pastré D. Bounedjah O, et al. Nucleic Acids Res. 2014 Jul;42(13):8678-91. doi: 10.1093/nar/gku582. Epub 2014 Jul 10. Nucleic Acids Res. 2014. PMID: 25013173 Free PMC article.
References
- Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5:417–421. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials