The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes - PubMed (original) (raw)
. 2007 May 8;104(19):7981-6.
doi: 10.1073/pnas.0611553104. Epub 2007 May 2.
Hang-Rae Kim, Jung-Hee Lee, Se-Yoon Kim, Jaejong Kim, Sunho Cha, Sang-Yoon Kim, Alistair C Darby, Hans-Henrik Fuxelius, Jun Yin, Ju Han Kim, Jihun Kim, Sang Joo Lee, Young-Sang Koh, Won-Jong Jang, Kyung-Hee Park, Siv G E Andersson, Myung-Sik Choi, Ik-Sang Kim
Affiliations
- PMID: 17483455
- PMCID: PMC1876558
- DOI: 10.1073/pnas.0611553104
The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes
Nam-Hyuk Cho et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Scrub typhus is caused by the obligate intracellular rickettsia Orientia tsutsugamushi (previously called Rickettsia tsutsugamushi). The bacterium is maternally inherited in trombicuid mites and transmitted to humans by feeding larvae. We report here the 2,127,051-bp genome of the Boryong strain, which represents the most highly repeated bacterial genome sequenced to date. The repeat density of the scrub typhus pathogen is 200-fold higher than that of its close relative Rickettsia prowazekii, the agent of epidemic typhus. A total of 359 tra genes for components of conjugative type IV secretion systems were identified at 79 sites in the genome. Associated with these are >200 genes for signaling and host-cell interaction proteins, such as histidine kinases, ankyrin-repeat proteins, and tetratrico peptide-repeat proteins. Additionally, the O. tsutsugamushi genome contains >400 transposases, 60 phage integrases, and 70 reverse transcriptases. Deletions and rearrangements have yielded unique gene combinations as well as frequent pseudogenization in the tra clusters. A comparative analysis of the tra clusters within the genome and across strains indicates sequence homogenization by gene conversion, whereas complexity, diversity, and pseudogenization are acquired by duplications, deletions, and transposon integrations into the amplified segments. The results suggest intragenomic duplications or multiple integrations of a massively proliferating conjugative transfer system. Diversifying selection on host-cell interaction genes along with repeated population bottlenecks may drive rare genome variants to fixation, thereby short-circuiting selection for low complexity in bacterial genomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
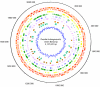
Fig. 1.
Circular map of the O. tsutsugamushi genome. The outer circle shows plus-strand genes and the second circle, minus-strand genes; in red, fragmented genes, and in orange, full-length genes. The third circle shows Rickettsial core genes in yellow; the fourth circle, identical repeats >200 bp in green; the fifth circle, tra genes for components of conjugative TFSSs in blue; the sixth circle, transposases in pink; in the seventh circle, numbered color blocks represent the tra gene clusters in Fig. 3; in the eighth circle, colored letters represent _tra_-associated gene clusters in Fig. 3; and the innermost circle shows the GC skew plot (G−C)/(G+C).
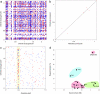
Fig. 2.
Repeat density in obligate intracellular bacteria. The frequency of repeats in the genomes of (a) O. tsutsugamushi and (b) R. prowazekii, with each dot representing a repeat >95% identical and >200 bp in size; blue, direct repeats; red, reverse repeats. (c) Scrambled gene order structures in O. tsutsugamushi and R. bellii. The yellow box indicates the location of the single tra cluster for TFSSs in the R. bellii genome and the homologous amplified tra clusters in the O. tsutsugamushi genomes. (d) Repeat density is plotted against genome size (% identical repeats >200 bp). Bacterial groupings are indicated by circles; red, obligate intracellular members of the Rickettsiales including Orientia, O. tsutsugamushi; Ana, Anaplasma marginale; Ehl, Ehrlichia ruminantium; Rb, R. bellii; Rc, Rickettsia conorii; Rf, R. felis; Rp, R. prowazekii; Rt, Rickettsia typhi; Trs, Wolbachia endosymbiont; TRS, wMel, Wolbachia wMel; green, Hel, Helicobacter pylori; Nei.c58, Neisseria meningitidis C58; Nei.z2491, N. meningitidis z2491; blue, the most highly repeated bacterial genomes until the time of this study are those of Myco, Mycoplasma mycoides; and Phyt, Phytoplasma strain OY M.
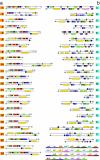
Fig. 3.
Gene order structure of (a) 24 long repeated tra gene clusters and (b) the flanking _tra_-associated gene clusters encoding putative effector proteins. The position of the clusters in the genome is indicated in Fig. 1.
Fig. 4.
A sequence comparison of selected O. tsutsugamushi repeated regions from different clinical isolates. (a) Expansion of region due to transposon insertion. (b) Reduction of the region due to deletion. The top line shows the gene order structure in the Boryong strain (B), and the bars underneath represent homologs regions found in strains Karp (K), Kato (KT), and Gilliam (G). Blue indicates inferred transposon insertions and red color sequence deletions. Green indicates the insertion of a transposon in strain G.
Similar articles
- Orientia tsutsugamushi Strain Ikeda Ankyrin Repeat-Containing Proteins Recruit SCF1 Ubiquitin Ligase Machinery via Poxvirus-Like F-Box Motifs.
Beyer AR, VieBrock L, Rodino KG, Miller DP, Tegels BK, Marconi RT, Carlyon JA. Beyer AR, et al. J Bacteriol. 2015 Oct;197(19):3097-109. doi: 10.1128/JB.00276-15. Epub 2015 Jul 13. J Bacteriol. 2015. PMID: 26170417 Free PMC article. - The Whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution.
Nakayama K, Yamashita A, Kurokawa K, Morimoto T, Ogawa M, Fukuhara M, Urakami H, Ohnishi M, Uchiyama I, Ogura Y, Ooka T, Oshima K, Tamura A, Hattori M, Hayashi T. Nakayama K, et al. DNA Res. 2008 Aug;15(4):185-99. doi: 10.1093/dnares/dsn011. Epub 2008 May 28. DNA Res. 2008. PMID: 18508905 Free PMC article. - Long-read whole genome sequencing and comparative analysis of six strains of the human pathogen Orientia tsutsugamushi.
Batty EM, Chaemchuen S, Blacksell S, Richards AL, Paris D, Bowden R, Chan C, Lachumanan R, Day N, Donnelly P, Chen S, Salje J. Batty EM, et al. PLoS Negl Trop Dis. 2018 Jun 6;12(6):e0006566. doi: 10.1371/journal.pntd.0006566. eCollection 2018 Jun. PLoS Negl Trop Dis. 2018. PMID: 29874223 Free PMC article. - New perspectives on rickettsial evolution from new genome sequences of rickettsia, particularly R. canadensis, and Orientia tsutsugamushi.
Eremeeva ME, Madan A, Shaw CD, Tang K, Dasch GA. Eremeeva ME, et al. Ann N Y Acad Sci. 2005 Dec;1063:47-63. doi: 10.1196/annals.1355.006. Ann N Y Acad Sci. 2005. PMID: 16481489 Review. - Orientia tsutsugamushi: An Unusual Intracellular Bacteria-Adaptation Strategies, Available Antibiotics, and Alternatives for Treatment.
Srivastava P, Shukla A, Singh R, Kant R, Mishra N, Behera SP, Dwivedi GR, Yadav DK. Srivastava P, et al. Curr Microbiol. 2024 Jun 21;81(8):236. doi: 10.1007/s00284-024-03754-1. Curr Microbiol. 2024. PMID: 38907107 Review.
Cited by
- Orientia tsutsugamushi subverts dendritic cell functions by escaping from autophagy and impairing their migration.
Choi JH, Cheong TC, Ha NY, Ko Y, Cho CH, Jeon JH, So I, Kim IK, Choi MS, Kim IS, Cho NH. Choi JH, et al. PLoS Negl Trop Dis. 2013;7(1):e1981. doi: 10.1371/journal.pntd.0001981. Epub 2013 Jan 3. PLoS Negl Trop Dis. 2013. PMID: 23301113 Free PMC article. - Repetitive DNA and next-generation sequencing: computational challenges and solutions.
Treangen TJ, Salzberg SL. Treangen TJ, et al. Nat Rev Genet. 2011 Nov 29;13(1):36-46. doi: 10.1038/nrg3117. Nat Rev Genet. 2011. PMID: 22124482 Free PMC article. Review. - Orientia tsutsugamushi Strain Ikeda Ankyrin Repeat-Containing Proteins Recruit SCF1 Ubiquitin Ligase Machinery via Poxvirus-Like F-Box Motifs.
Beyer AR, VieBrock L, Rodino KG, Miller DP, Tegels BK, Marconi RT, Carlyon JA. Beyer AR, et al. J Bacteriol. 2015 Oct;197(19):3097-109. doi: 10.1128/JB.00276-15. Epub 2015 Jul 13. J Bacteriol. 2015. PMID: 26170417 Free PMC article. - Anaplasma phagocytophilum and Ehrlichia chaffeensis type IV secretion and Ank proteins.
Rikihisa Y, Lin M. Rikihisa Y, et al. Curr Opin Microbiol. 2010 Feb;13(1):59-66. doi: 10.1016/j.mib.2009.12.008. Epub 2010 Jan 5. Curr Opin Microbiol. 2010. PMID: 20053580 Free PMC article. Review. - Gene expression and involvement of signaling pathways during host-pathogen interplay in Orientia tsutsugamushi infection.
Panda S, Swain SK, Sahu BP, Sarangi R. Panda S, et al. 3 Biotech. 2022 Sep;12(9):180. doi: 10.1007/s13205-022-03239-7. Epub 2022 Jul 19. 3 Biotech. 2022. PMID: 35860421 Free PMC article. Review.
References
- Seong SY, Choi MS, Kim IS. Microbes Infect. 2001;3:11–21. - PubMed
- Walker DH. Biology of Rickettsial Disease. Boca Raton, FL: CRC; 1998.
- Kawamura A, Tanaka H, Tamura A. Tsutsugamushi Disease. Tokyo, Japan: University of Tokyo Press; 1995.
- Philip CB. J Parasitol. 1948;34:169–191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases