PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations - PubMed (original) (raw)
. 2007 Jul;35(Web Server issue):W522-5.
doi: 10.1093/nar/gkm276. Epub 2007 May 8.
Affiliations
- PMID: 17488841
- PMCID: PMC1933214
- DOI: 10.1093/nar/gkm276
PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations
Todd J Dolinsky et al. Nucleic Acids Res. 2007 Jul.
Abstract
Real-world observable physical and chemical characteristics are increasingly being calculated from the 3D structures of biomolecules. Methods for calculating pK(a) values, binding constants of ligands, and changes in protein stability are readily available, but often the limiting step in computational biology is the conversion of PDB structures into formats ready for use with biomolecular simulation software. The continued sophistication and integration of biomolecular simulation methods for systems- and genome-wide studies requires a fast, robust, physically realistic and standardized protocol for preparing macromolecular structures for biophysical algorithms. As described previously, the PDB2PQR web server addresses this need for electrostatic field calculations (Dolinsky et al., Nucleic Acids Research, 32, W665-W667, 2004). Here we report the significantly expanded PDB2PQR that includes the following features: robust standalone command line support, improved pK(a) estimation via the PROPKA framework, ligand parameterization via PEOE_PB charge methodology, expanded set of force fields and easily incorporated user-defined parameters via XML input files, and improvement of atom addition and optimization code. These features are available through a new web interface (http://pdb2pqr.sourceforge.net/), which offers users a wide range of options for PDB file conversion, modification and parameterization.
Figures
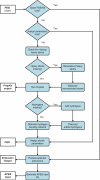
Figure 1.
Flowchart demonstrating the sequence of operations performed by the pipeline. The process begins with an input PDB file and ends with a parameterized PQR file and, optionally, an APBS input file.
Similar articles
- PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations.
Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. Dolinsky TJ, et al. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W665-7. doi: 10.1093/nar/gkh381. Nucleic Acids Res. 2004. PMID: 15215472 Free PMC article. - Web servers and services for electrostatics calculations with APBS and PDB2PQR.
Unni S, Huang Y, Hanson RM, Tobias M, Krishnan S, Li WW, Nielsen JE, Baker NA. Unni S, et al. J Comput Chem. 2011 May;32(7):1488-91. doi: 10.1002/jcc.21720. Epub 2011 Feb 1. J Comput Chem. 2011. PMID: 21425296 Free PMC article. - Patch Finder Plus (PFplus): a web server for extracting and displaying positive electrostatic patches on protein surfaces.
Shazman S, Celniker G, Haber O, Glaser F, Mandel-Gutfreund Y. Shazman S, et al. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W526-30. doi: 10.1093/nar/gkm401. Epub 2007 May 30. Nucleic Acids Res. 2007. PMID: 17537808 Free PMC article. - CHARMM-GUI 10 years for biomolecular modeling and simulation.
Jo S, Cheng X, Lee J, Kim S, Park SJ, Patel DS, Beaven AH, Lee KI, Rui H, Park S, Lee HS, Roux B, MacKerell AD Jr, Klauda JB, Qi Y, Im W. Jo S, et al. J Comput Chem. 2017 Jun 5;38(15):1114-1124. doi: 10.1002/jcc.24660. Epub 2016 Nov 14. J Comput Chem. 2017. PMID: 27862047 Free PMC article. Review. - Biomolecular force fields: where have we been, where are we now, where do we need to go and how do we get there?
Dauber-Osguthorpe P, Hagler AT. Dauber-Osguthorpe P, et al. J Comput Aided Mol Des. 2019 Feb;33(2):133-203. doi: 10.1007/s10822-018-0111-4. Epub 2018 Nov 30. J Comput Aided Mol Des. 2019. PMID: 30506158 Review.
Cited by
- Two crystal structures of Bombyx mori lipoprotein 3 - structural characterization of a new 30-kDa lipoprotein family member.
Pietrzyk AJ, Bujacz A, Mueller-Dieckmann J, Lochynska M, Jaskolski M, Bujacz G. Pietrzyk AJ, et al. PLoS One. 2013 Apr 16;8(4):e61303. doi: 10.1371/journal.pone.0061303. Print 2013. PLoS One. 2013. PMID: 23613829 Free PMC article. - Substrate channeling between the human dihydrofolate reductase and thymidylate synthase.
Wang N, McCammon JA. Wang N, et al. Protein Sci. 2016 Jan;25(1):79-86. doi: 10.1002/pro.2720. Epub 2015 Jun 29. Protein Sci. 2016. PMID: 26096018 Free PMC article. - Dynamic regulation of Zn(II) sequestration by calgranulin C.
Wang Q, Kuci D, Bhattacharya S, Hadden-Perilla JA, Gupta R. Wang Q, et al. Protein Sci. 2022 Sep;31(9):e4403. doi: 10.1002/pro.4403. Protein Sci. 2022. PMID: 36367084 Free PMC article. - Modeling of interaction between cytochrome c and the WD domains of Apaf-1: bifurcated salt bridges underlying apoptosome assembly.
Shalaeva DN, Dibrova DV, Galperin MY, Mulkidjanian AY. Shalaeva DN, et al. Biol Direct. 2015 May 27;10:29. doi: 10.1186/s13062-015-0059-4. Biol Direct. 2015. PMID: 26014357 Free PMC article. - Differential hERG ion channel activity of ultrasmall gold nanoparticles.
Leifert A, Pan Y, Kinkeldey A, Schiefer F, Setzler J, Scheel O, Lichtenbeld H, Schmid G, Wenzel W, Jahnen-Dechent W, Simon U. Leifert A, et al. Proc Natl Acad Sci U S A. 2013 May 14;110(20):8004-9. doi: 10.1073/pnas.1220143110. Epub 2013 Apr 29. Proc Natl Acad Sci U S A. 2013. PMID: 23630249 Free PMC article.
References
- Baker NA. Improving implicit solvent simulations: a Poisson-centric view. Curr. Opin. Struct. Biol. 2005;15:137–143. - PubMed
- Darden TA. In: Computational Biochemistry and Biophysics. Becker OM, MacKerell AD Jr, Roux B, Watanabe M, editors. New York: Marcel Dekker Inc; 2001. pp. 91–114.
- Roux B. In: Computational Biochemistry and Biophysics. Becker OM, MacKerell AD Jr, Roux B, Watanabe M, editors. New York: Marcel Dekker; 2001. pp. 133–152.
- Davis ME, McCammon JA. Electrostatics in biomolecular structure and dynamics. Chem. Rev. 1990;94:7684–7692.
- Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM069702-04/GM/NIGMS NIH HHS/United States
- R01 GM069702-03/GM/NIGMS NIH HHS/United States
- P41 RR08605/RR/NCRR NIH HHS/United States
- GM069702/GM/NIGMS NIH HHS/United States
- R01 GM069702-01/GM/NIGMS NIH HHS/United States
- P41 RR008605/RR/NCRR NIH HHS/United States
- R01 GM069702/GM/NIGMS NIH HHS/United States
- R01 GM069702-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials