VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease - PubMed (original) (raw)
VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease
Dayle A Daines et al. J Bacteriol. 2007 Jul.
Abstract
Nontypeable Haemophilus influenzae (NTHi) organisms are obligate parasites of the human upper respiratory tract that can exist as commensals or pathogens. Toxin-antitoxin (TA) loci are highly conserved gene pairs that encode both a toxin and antitoxin moiety. Seven TA gene families have been identified to date, and NTHi carries two alleles of the vapBC family. Here, we have characterized the function of one of the NTHi alleles, vapBC-1. The gene pair is transcribed as an operon in two NTHi clinical isolates, and promoter fusions display an inverse relationship to culture density. The antitoxin VapB-1 forms homomultimers both in vitro and in vivo. The expression of the toxin VapC-1 conferred growth inhibition to an Escherichia coli expression strain and was successfully purified only when cloned in tandem with its cognate antitoxin. Using total RNA isolated from both E. coli and NTHi, we show for the first time that VapC-1 is an RNase that is active on free RNA but does not degrade DNA in vitro. Preincubation of the purified toxin and antitoxin together results in the formation of a protein complex that abrogates the activity of the toxin. We conclude that the NTHi vapBC-1 gene pair functions as a classical TA locus and that the induction of VapC-1 RNase activity leads to growth inhibition via the mechanism of mRNA cleavage.
Figures
FIG. 1.
NTHi strains R2866 and 86-028NP express vapBC-1 as an operon. Reverse transcriptase PCRs with a forward primer in vapB-1 and a reverse primer in vapC-1 electrophoresed on a 0.8% agarose gel. Lanes 1 and 4, 1-kb DNA ladder; lanes 2 and 5, R2866 and 86-028NP with reverse transcriptase (+RT); lanes 3 and 6, R2866 and 86-028NP without reverse transcriptase (−RT).
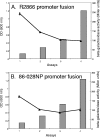
FIG. 2.
Expression of vapBC-1 over the cell cycle. (A) The β-galactosidase activity of the R2866 vapBC-1 promoter fusion (curve) is inversely related to culture density (bars). (B) The 86-028NP vapBC-1 promoter fusion (curve) displays lower initial β-galactosidase activity than the R2866 fusion but mirrors the trend toward decreasing expression with increasing culture density (bars). OD, optical density.
FIG. 3.
VapB-1 forms multimers in vitro. (A) Coomassie-stained 12% SDS-PAGE separation of a typical purified pTrc::VapB-1 construct (2 μg) from DH5α. (B) An identical immunoblot probed with anti-Xpress-HRP monoclonal antibody. Note the multiple bands resulting from apparent homo-interactions.
FIG. 4.
VapB-1 homodimerizes in vivo. With no protein fused to the LexA DBD, the repressor cannot form a dimer, and transcription of the lacZ reporter gene is constitutive (gray bar). However, when the LexA DBD is fused to VapB-1, competent dimers are formed, and the chimeras can bind to the LexA operator sequence, repressing transcription of the reporter gene (black bar). This level of repression indicates strong VapB-1 homo-interaction.
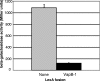
FIG. 5.
VapC-1 causes growth inhibition in vivo. When grown in LB broth without IPTG, the pTrcHisA vector promoter allowed a small amount of transcription of pTrc::VapC-1, which conferred growth inhibition to the DH5α expression strain and made this construct unsuitable for protein isolation. No significant growth effects were observed with the pTrc::VapB-1 fusion or the vector alone. OD, optical density.
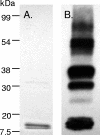
FIG. 6.
VapC-1 was successfully purified via tandem cloning into pET24b and expression in BL21(DE3). (A) The vapBC-1 operon was fused to pET24b such that vapB-1 was in frame with the vector's ATG start codon at the N terminus, and vapC-1 was in frame with the C-terminal polyhistidine tag, creating pDD686. Induction of the construct with IPTG resulted in no significant growth inhibition of the expression strain. (B) Coomassie-stained 12% SDS-PAGE separation of a typical purified pET::VapC-1 construct (3.5 μg). Two bands are visible: one at the calculated molecular mass of pET::VapC-1 (16.6 kDa) and one at the size of pET::VapB-1 (10.5 kDa). (C) Identical immunoblot probed with anti-His C-terminal HRP-linked monoclonal antibody. Note that only a single band is apparent.
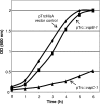
FIG. 7.
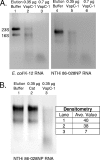
VapC-1 is an RNase toxin. (A) Total RNA from E. coli K-12 or H. influenzae strain 86-028NP was used as the substrate in RNase activity assays with increasing amounts of the purified VapC-1 toxin. Lanes 1 and 4, MagneHis protein elution buffer control; lanes 2 and 5, 0.35 μg of VapC-1; lanes 3 and 6, 0.7 μg of VapC-1. (B) The Cat protein was cloned into pET24b and purified in the same manner as VapC-1 as a control for any copurifying RNase activity. Lane 1, MagneHis protein elution buffer control; lane 2, 0.35 μg of Cat protein; lane 3, 0.35 μg of VapC-1. Densitometry indicated that the observed RNase activity was specific to VapC-1. Ave, average.
FIG. 8.
VapB-1 forms nontoxic complexes with VapC-1 in vitro. The antitoxin VapB-1 was cloned into pET24b as a single gene and purified using the MagneHis native protein purification protocol. Various amounts of purified VapB-1 were incubated with a constant amount of VapC-1 for 30 min prior to the addition of the total RNA substrate in RNase activity assays. A 4:1 ratio of VapB-1 to VapC-1 abrogates the RNase activity of VapC-1. Lanes 1 and 6, MagneHis protein elution buffer control; lanes 2 and 7, 0.2 μg of VapB-1; lanes 3 and 8, 0.4 μg of VapB-1 plus 0.1 μg of VapC-1 (4:1 ratio); lanes 4 and 9, 0.2 μg of VapB-1 plus 0.1 μg of VapC-1 (2:1 ratio); lanes 5 and 10, 0.1 μg of VapC-1 alone. For these assays, H. influenzae strain R2866 total RNA was used. Note the natural 23S fragmentation pattern, which differs from that of strain 86-028NP.
Similar articles
- Crystal Structure of VapBC-1 from Nontypeable Haemophilus influenzae and the Effect of PIN Domain Mutations on Survival during Infection.
Molinaro AL, Kashipathy MM, Lovell S, Battaile KP, Coussens NP, Shen M, Daines DA. Molinaro AL, et al. J Bacteriol. 2019 May 22;201(12):e00026-19. doi: 10.1128/JB.00026-19. Print 2019 Jun 15. J Bacteriol. 2019. PMID: 30936373 Free PMC article. - Homologous VapC Toxins Inhibit Translation and Cell Growth by Sequence-Specific Cleavage of tRNAfMet.
Walling LR, Butler JS. Walling LR, et al. J Bacteriol. 2018 Jan 10;200(3):e00582-17. doi: 10.1128/JB.00582-17. Print 2018 Feb 1. J Bacteriol. 2018. PMID: 29109187 Free PMC article. - Toxin-antitoxin loci vapBC-1 and vapXD contribute to survival and virulence in nontypeable Haemophilus influenzae.
Ren D, Walker AN, Daines DA. Ren D, et al. BMC Microbiol. 2012 Nov 19;12:263. doi: 10.1186/1471-2180-12-263. BMC Microbiol. 2012. PMID: 23157645 Free PMC article. - The PIN-domain ribonucleases and the prokaryotic VapBC toxin-antitoxin array.
Arcus VL, McKenzie JL, Robson J, Cook GM. Arcus VL, et al. Protein Eng Des Sel. 2011 Jan;24(1-2):33-40. doi: 10.1093/protein/gzq081. Epub 2010 Oct 29. Protein Eng Des Sel. 2011. PMID: 21036780 Review. - Cut to the chase--Regulating translation through RNA cleavage.
Sofos N, Xu K, Dedic E, Brodersen DE. Sofos N, et al. Biochimie. 2015 Jul;114:10-7. doi: 10.1016/j.biochi.2015.01.009. Epub 2015 Jan 27. Biochimie. 2015. PMID: 25633441 Review.
Cited by
- VapC6, a ribonucleolytic toxin regulates thermophilicity in the crenarchaeote Sulfolobus solfataricus.
Maezato Y, Daugherty A, Dana K, Soo E, Cooper C, Tachdjian S, Kelly RM, Blum P. Maezato Y, et al. RNA. 2011 Jul;17(7):1381-92. doi: 10.1261/rna.2679911. Epub 2011 May 27. RNA. 2011. PMID: 21622901 Free PMC article. - Analysis of non-typeable Haemophilous influenzae VapC1 mutations reveals structural features required for toxicity and flexibility in the active site.
Hamilton B, Manzella A, Schmidt K, DiMarco V, Butler JS. Hamilton B, et al. PLoS One. 2014 Nov 12;9(11):e112921. doi: 10.1371/journal.pone.0112921. eCollection 2014. PLoS One. 2014. PMID: 25391136 Free PMC article. - Growth and translation inhibition through sequence-specific RNA binding by Mycobacterium tuberculosis VapC toxin.
Sharp JD, Cruz JW, Raman S, Inouye M, Husson RN, Woychik NA. Sharp JD, et al. J Biol Chem. 2012 Apr 13;287(16):12835-47. doi: 10.1074/jbc.M112.340109. Epub 2012 Feb 21. J Biol Chem. 2012. PMID: 22354968 Free PMC article. - Regulation of the vapBC-1 toxin-antitoxin locus in nontypeable Haemophilus influenzae.
Cline SD, Saleem S, Daines DA. Cline SD, et al. PLoS One. 2012;7(3):e32199. doi: 10.1371/journal.pone.0032199. Epub 2012 Mar 13. PLoS One. 2012. PMID: 22427824 Free PMC article. - Discovery of Small-Molecule VapC1 Nuclease Inhibitors by Virtual Screening and Scaffold Hopping from an Atomic Structure Revealing Protein-Protein Interactions with a Native VapB1 Inhibitor.
Sun H, Coussens NP, Danchik C, Wachsmuth LM, Henderson MJ, Patnaik S, Hall MD, Molinaro AL, Daines DA, Shen M. Sun H, et al. J Chem Inf Model. 2022 Mar 14;62(5):1249-1258. doi: 10.1021/acs.jcim.1c01188. Epub 2022 Feb 1. J Chem Inf Model. 2022. PMID: 35103473 Free PMC article.
References
- Arandjus, C., P. N. Black, P. J. Poole, R. W. Baker, and C. Steurer-Stey. 2006. Oral bacterial vaccines for the prevention of acute exacerbations in chronic obstructive pulmonary disease and chronic bronchitis. Respir. Med. 100:1671-1681. - PubMed
- Arcus, V. L., P. B. Rainey, and S. J. Turner. 2005. The PIN-domain toxin-antitoxin array in mycobacteria. TRENDS Microbiol. 13:360-365. - PubMed
- Casadaban, M. J., J. Chou, and S. N. Cohen. 1980. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J. Bacteriol. 143:971-980. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases