Huntingtin interacting proteins are genetic modifiers of neurodegeneration - PubMed (original) (raw)
doi: 10.1371/journal.pgen.0030082.
Eliana Romero, Robert R Becklin, Rakesh Chettier, Russell Bell, Amit Phansalkar, Andrew Strand, Cameron Torcassi, Justin Savage, Anthony Hurlburt, Guang-Ho Cha, Lubna Ukani, Cindy Lou Chepanoske, Yuejun Zhen, Sudhir Sahasrabudhe, James Olson, Cornelia Kurschner, Lisa M Ellerby, John M Peltier, Juan Botas, Robert E Hughes
Affiliations
- PMID: 17500595
- PMCID: PMC1866352
- DOI: 10.1371/journal.pgen.0030082
Huntingtin interacting proteins are genetic modifiers of neurodegeneration
Linda S Kaltenbach et al. PLoS Genet. 2007.
Abstract
Huntington's disease (HD) is a fatal neurodegenerative condition caused by expansion of the polyglutamine tract in the huntingtin (Htt) protein. Neuronal toxicity in HD is thought to be, at least in part, a consequence of protein interactions involving mutant Htt. We therefore hypothesized that genetic modifiers of HD neurodegeneration should be enriched among Htt protein interactors. To test this idea, we identified a comprehensive set of Htt interactors using two complementary approaches: high-throughput yeast two-hybrid screening and affinity pull down followed by mass spectrometry. This effort led to the identification of 234 high-confidence Htt-associated proteins, 104 of which were found with the yeast method and 130 with the pull downs. We then tested an arbitrary set of 60 genes encoding interacting proteins for their ability to behave as genetic modifiers of neurodegeneration in a Drosophila model of HD. This high-content validation assay showed that 27 of 60 orthologs tested were high-confidence genetic modifiers, as modification was observed with more than one allele. The 45% hit rate for genetic modifiers seen among the interactors is an order of magnitude higher than the 1%-4% typically observed in unbiased genetic screens. Genetic modifiers were similarly represented among proteins discovered using yeast two-hybrid and pull-down/mass spectrometry methods, supporting the notion that these complementary technologies are equally useful in identifying biologically relevant proteins. Interacting proteins confirmed as modifiers of the neurodegeneration phenotype represent a diverse array of biological functions, including synaptic transmission, cytoskeletal organization, signal transduction, and transcription. Among the modifiers were 17 loss-of-function suppressors of neurodegeneration, which can be considered potential targets for therapeutic intervention. Finally, we show that seven interacting proteins from among 11 tested were able to co-immunoprecipitate with full-length Htt from mouse brain. These studies demonstrate that high-throughput screening for protein interactions combined with genetic validation in a model organism is a powerful approach for identifying novel candidate modifiers of polyglutamine toxicity.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
Figure 1. Results of the Physical Interaction Screens
(A) Overview of the discovery workflow is represented. Y2H and pull-down/MS workflows are shown on the left and right, respectively. The number of Htt-fragment baits used for Y2H searches or pull downs includes wild-type (23 Q) and mutant (48 Q and 55 Q) forms. Not all Htt fragments were successfully expressed in bacteria or yielded positive interactions in Y2H screens. For Y2H positive prey identification, the top blast score was chosen. The total number of genes found in pull down/MS includes 15 mouse and human homologs; the nonredundant set does not include mouse homologs (see Supporting Information). (B) A diagram of Htt baits used in Y2H and MS experiments is presented. Structural features (HEAT repeat domains and protease cleavage region) are indicated by shaded boxes on a diagram of the Htt protein. Numbers indicate reference amino acids positions (with respect to NP_002102). Lines representing Htt-fragment baits with associated amino acid positions indicated by numbers are shown relative to the diagram of Htt. We purified three Htt-fragments (top panel) from bacteria in sufficient quantities for pull-down/MS experiments. Htt-fragment baits used in Y2H screens are shown on the bottom panel. Some baits did not yield positive interactions (dotted lines). Htt clones that contained the polyQ sequence were generated in wild-type (23 Q) and expanded (55 Q, 75 Q, and 97 Q) forms (asterisk). (C) A functional analysis of Htt-fragment-interacting proteins is presented. The number of proteins representing the indicated functional categories found in Htt-fragment Y2H screens (white bars) or pull down/MS (black bars) are shown. Proteins were assigned to categories based on gene ontology. Only categories with more than one protein assigned are shown.
Figure 2. Modification of the Phenotypes Caused by N-Terminal Expanded Htt in the Drosophila Eye
Retinal sections of adult Drosophila eyes show modification of the phenotypes caused by expression of different levels (B and I) of a transgene encoding an N-terminal expanded Htt fragment. Enhancers (C–G) and suppressors (J–N) include proteins involved in cytoskeletal organization (C) and (J), signal transduction (D) and (K), neurotransmitter secretion (E) and (L), proteolysis/peptidolysis and the ubiquitin cycle (F) and (M), and transcriptional/translational regulation (G) and (N). Retinal sections of day 5 control flies cultured at 25 °C expressing the gene that encodes expanded N-terminal Htt fragment (GMR-GAL4/+; UAS:128Qhtt[M64]/+) (B) show a degenerative phenotype when compared to controls of the same age and cultured at the same temperature (GMR-GAL4/+) (A). The phenotype consists of a shortening (see arrow) and detachment of the retina, as well as the presence of vacuoles in the retina. The Htt-fragment-induced phenotype can be enhanced by (C) reduced levels of zipper (GMR-GAL4/P{PZ}zip02957; UAS:128Qhtt[M64]/+), (D) reduced levels of Src oncogene at 42A (GMR-GAL4/P{lacW}Src42Ak10108; UAS:128Qhtt[M64]/+), (E) overexpression of soluble N-ethylmaleimide-sensitive -attachment protein (GMR-GAL4/+; UAS:128Qhtt[M64]/UAS-S102C#2D_),_ (F) reduced levels of fat facets (GMR-GAL4/+; UAS:128Qhtt[M64]/fafBx4 ), and (G) reduced levels of crooked legs (GMR-GAL4/P{PZ}crol04418cn1; UAS:128Qhtt[M64]/+). None of these mutations cause an abnormal eye phenotype in flies carrying the GMR-GAL4 driver but not the UAS:128Qhtt[M64] transgene (unpublished data). However, when combined with an N-terminal expanded Htt fragment, they lead to an even larger decrease in retinal thickness sometimes accompanied by an increase in retinal detachment and vacuolization. Retinal sections of day 1 control flies cultured at 27 °C expressing a gene that encodes an expanded N-terminal Htt fragment (GMR-GAL4/+; UAS:128Qhtt[M64]/+) (I) show a severe degenerative phenotype when compared to GMR controls of the same age and cultured at the same temperature (H). The phenotype consists of a shortening (see arrow) and detachment of the retina, as well as the presence of vacuoles in the retina. The Htt-fragment-induced phenotype can be suppressed by (J) reduced levels of hu li tai shao (GMR-GAL4/P{lacW}htsk06121; UAS:128Qhtt[M64]/+), (K) reduced levels of G protein iαsubunit 65A (GMR-GAL4/; UAS:128Qhtt[M64]/P{SUPor-P}G-ia65AKG01907ry506 ), (L) reduced levels of clathrin heavy chain (Chc1/+ GMR-GAL4/+; UAS:128Qhtt[M64]/+), (M) reduced levels of Rpt1 (GMR-GAL4/P{PZ}Rpt105643cn1; UAS:128Qhtt[M64]/+), and (N) reduced levels of myocyte enhancing factor 2 (GMR-GAL4/Df(2R)X1,Mef2[X1]; UAS:128Qhtt[M64]/+). These mutations decrease the vacuolization and increase the retinal thickness as well as virtually eliminating the retinal detachment.
Figure 3. Modification of the Expanded Htt-Fragment-Induced Phenotype by a STX1A Loss-of-Function Mutation
Modification was observed both in the eye (external phenotype and retinal sections) and in the nervous system (climbing ability and survival). (A) Retinal sections of day 5 flies raised at 25 °C (left), day 1 flies raised at 27 °C (middle), and standard error of mean of day 5 flies raised at 29 °C (right) expressing a gene that encodes expanded N-terminal Htt fragment (GMR-GAL4/+; UAS:128Qhtt[M64]/+). (B) Retinal sections of day 5 flies raised at 25 °C (left), day 1 flies raised at 27 °C (middle), and standard error of mean of day 5 flies raised at 29 °C (right) expressing a gene that encodes expanded N-terminal Htt fragment and carrying reduced levels of STX1A (GMR-GAL4/+; UAS:128Qhtt[M64]/Syx1A229ry506). Note suppression of both the retinal and external eye phenotypes at all three temperatures. Overexpression of STX1A shows enhancement of the retinal degeneration and external 128 Qhtt phenotype (unpublished data). (C) Climbing assay (top) and survival assay (bottom) results confirm the suppression observed in the eye assay. Shown in red/pink are the climbing performance and survival curve of a population of females flies expressing a gene that encodes expanded N-terminal expanded Htt fragment (elav-GAL4/+; UAS:128Qhtt[F27B]/+). Shown in blue/light blue are the improved climbing performance and survival curve of a population of females flies expressing a gene that encodes expanded N-terminal Htt fragment and carrying a heterozygous loss-of-function mutation in STX1A (elav-GAL4/+; +/+; UAS:128Qhtt[F27B]/Syx1A229 ry506). (x-Axis, age of flies in days; y-axis, percent surviving or climbing flies; LOF, loss-of-function).
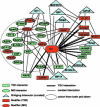
Figure 4. A Network of Protein Interactions Involved in Vesicle Traffic
A network is shown that includes protein interactions described in this study and interactions curated from the public domain (NCBI Entrez Gene). Htt-fragment-interacting proteinss found in this study are indicated as ovals (MS) or rectangles (Y2H). Binary Y2H interactions found in this study are indicated as thick lines. Proteins contained in the dotted circle were identified in Htt-fragment pull downs using brain lysates. Thin lines indicate curated protein interactions. Curated bridging interactions (blue triangles) are defined as proteins reported to interact with HD and at least one other protein in the network. Proteins whose Drosophila ortholog genes acted as modifiers in this study are indicated in red.
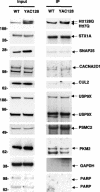
Figure 5. Co-Immunoprecipitation of Huntingtin-Interacting Proteins from YAC128 Mouse Brain
Htt was immunoprecipitated with mouse monoclonal Htt antibody and probed with rabbit polyclonal Htt BKP1 antibody (top right panel). The input for each protein (left panels) and resulting immunoprecipitation are shown (right panels). The lower molecular weight band in the PKM2 immunoprecipitation is an immunoglobulin (IgG) band. GAPDH is included as a positive control. PARP is included as a negative control.
Similar articles
- Adenovirus vector-based in vitro neuronal cell model for Huntington's disease with human disease-like differential aggregation and degeneration.
Dong X, Zong S, Witting A, Lindenberg KS, Kochanek S, Huang B. Dong X, et al. J Gene Med. 2012 Jul;14(7):468-81. doi: 10.1002/jgm.2641. J Gene Med. 2012. PMID: 22700462 - Mutant Huntingtin Protein Interaction Map Implicates Dysregulation of Multiple Cellular Pathways in Neurodegeneration of Huntington's Disease.
Podvin S, Rosenthal SB, Poon W, Wei E, Fisch KM, Hook V. Podvin S, et al. J Huntingtons Dis. 2022;11(3):243-267. doi: 10.3233/JHD-220538. J Huntingtons Dis. 2022. PMID: 35871359 Free PMC article. - Selective degeneration in YAC mouse models of Huntington disease.
Van Raamsdonk JM, Warby SC, Hayden MR. Van Raamsdonk JM, et al. Brain Res Bull. 2007 Apr 30;72(2-3):124-31. doi: 10.1016/j.brainresbull.2006.10.018. Epub 2006 Nov 16. Brain Res Bull. 2007. PMID: 17352936 Review. - Huntingtin affinity for partners is not changed by polyglutamine length: aggregation itself triggers aberrant interactions.
Davranche A, Aviolat H, Zeder-Lutz G, Busso D, Altschuh D, Trottier Y, Klein FA. Davranche A, et al. Hum Mol Genet. 2011 Jul 15;20(14):2795-806. doi: 10.1093/hmg/ddr178. Epub 2011 Apr 25. Hum Mol Genet. 2011. PMID: 21518730 - Huntingtin and its role in neuronal degeneration.
Li SH, Li XJ. Li SH, et al. Neuroscientist. 2004 Oct;10(5):467-75. doi: 10.1177/1073858404266777. Neuroscientist. 2004. PMID: 15359012 Review.
Cited by
- Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis.
Ratovitski T, Chighladze E, Arbez N, Boronina T, Herbrich S, Cole RN, Ross CA. Ratovitski T, et al. Cell Cycle. 2012 May 15;11(10):2006-21. doi: 10.4161/cc.20423. Epub 2012 May 15. Cell Cycle. 2012. PMID: 22580459 Free PMC article. - HTT (huntingtin) and RAB7 co-migrate retrogradely on a signaling LAMP1-containing late endosome during axonal injury.
Krzystek TJ, White JA, Rathnayake R, Thurston L, Hoffmar-Glennon H, Li Y, Gunawardena S. Krzystek TJ, et al. Autophagy. 2023 Apr;19(4):1199-1220. doi: 10.1080/15548627.2022.2119351. Epub 2022 Sep 9. Autophagy. 2023. PMID: 36048753 Free PMC article. - Taming the Huntington's Disease Proteome: What Have We Learned?
Seeley C, Kegel-Gleason KB. Seeley C, et al. J Huntingtons Dis. 2021;10(2):239-257. doi: 10.3233/JHD-200465. J Huntingtons Dis. 2021. PMID: 33998547 Free PMC article. Review. - Large-scale functional RNAi screen in C. elegans identifies genes that regulate the dysfunction of mutant polyglutamine neurons.
Lejeune FX, Mesrob L, Parmentier F, Bicep C, Vazquez-Manrique RP, Parker JA, Vert JP, Tourette C, Neri C. Lejeune FX, et al. BMC Genomics. 2012 Mar 13;13:91. doi: 10.1186/1471-2164-13-91. BMC Genomics. 2012. PMID: 22413862 Free PMC article. - Advances in the understanding of nuclear pore complexes in human diseases.
Li Y, Zhu J, Zhai F, Kong L, Li H, Jin X. Li Y, et al. J Cancer Res Clin Oncol. 2024 Jul 30;150(7):374. doi: 10.1007/s00432-024-05881-5. J Cancer Res Clin Oncol. 2024. PMID: 39080077 Free PMC article. Review.
References
- Hickey MA, Chesselet MF. The use of transgenic and knock-in mice to study Huntington's disease. Cytogenet Genome Res. 2003;100:276–286. - PubMed
- Zoghbi HY, Botas J. Mouse and fly models of neurodegeneration. Trends Genet. 2002;18:463–471. - PubMed
- Driscoll M, Gerstbrein B. Dying for a cause: Invertebrate genetics takes on human neurodegeneration. Nat Rev Genet. 2003;4:181–194. - PubMed
- DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS40251/NS/NINDS NIH HHS/United States
- R01 NS042179/NS/NINDS NIH HHS/United States
- R56 NS042179/NS/NINDS NIH HHS/United States
- NS42179/NS/NINDS NIH HHS/United States
- R01 NS040251/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials