Differential regulation of NF-kappaB by elongation factors is determined by core promoter type - PubMed (original) (raw)
Differential regulation of NF-kappaB by elongation factors is determined by core promoter type
Liat Amir-Zilberstein et al. Mol Cell Biol. 2007 Jul.
Abstract
NF-kappaB transcription factors activate genes important for immune response, inflammation, and cell survival. P-TEFb and DSIF, which are positive and negative transcription elongation factors, respectively, both regulate NF-kappaB-induced transcription, but the mechanism underlying their recruitment to NF-kappaB target genes is unknown. We show here that upon induction of NF-kappaB, a subset of target genes is regulated differentially by either P-TEFb or DSIF. The regulation of these genes and their occupancy by these elongation factors are dependent on the NF-kappaB enhancer and the core promoter type. Converting a TATA-less promoter to a TATA promoter switches the regulation of NF-kappaB from DSIF to P-TEFb. Accumulation or displacement of DSIF and P-TEFb is dictated by the formation of distinct initiation complexes (TFIID dependent or independent) on the two types of core promoter. The underlying mechanism for the dissociation of DSIF from TATA promoters upon NF-kappaB activation involves the phosphorylation of RNA polymerase II by P-TEFb. The results highlight a regulatory link between the initiation and the elongation phases of the transcription reaction and broaden our comprehension of the NF-kappaB pathway.
Figures
FIG. 1.
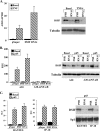
Attenuation of NF-κB-mediated transcription by DSIF. (A) 293T cells were transfected with either pSuper or DSIF p160 RNAi, and 48 h posttransfection, cells were treated with TNF-α for 1 h. Total RNA was extracted and subjected to a quantitative RT-PCR for the A20 and GAPDH mRNAs, using a Light Cycler system. Data are means and standard deviations of three independent experiments. A representative immunoblot verifying down regulation of p160 DSIF is shown on the right. (B) 293T cells were transfected with either the wild-type A20 or the A20-mNF-κB (in which the two NF-κB sites are mutated) promoter, with or without the p65/RelA (p65) expression vector, and with either pSuper (parental vector) or DSIF p160 RNAi as indicated. Cells were harvested 48 h posttransfection, and luciferase activity was measured. RSV promoter-driven Renilla luciferase reporter plasmids served to normalize the transfection efficiency. Shown is the relative luciferase activity (luciferase units divided by the activity of cotransfected RSV promoter-driven Renilla reporter luciferase, in relative light units [RLU]). Data are means and standard deviations of seven independent experiments, each with independent duplicates (left). A representative immunoblot showing DSIF knockdown in transfected cells is shown on the right. (C) 293T cells were cotransfected with p65/RelA expression plasmid together with luciferase reporter genes driven by the RANTES and IP-10 promoters and with either pSuper or DSIF p160 RNAi as indicated. The averages of seven independent transfection experiments are shown in the graphs. A representative immunoblot showing DSIF knockdown in cells transfected with the indicated reporter plasmids together with p65/RelA is shown on the right.
FIG. 2.
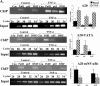
DSIF attenuation of NF-κB is dependent on a TATA-less core promoter. (A) 293T cells were cotransfected with A20 promoter mutants (schematically shown on the right) with a suboptimal concentration of p65/RelA expression vector and with either pSuper (parental vector) or DSIF p160 RNAi as indicated and analyzed as described for Fig. 1B. The effect of DSIF knockdown on different A20 promoter mutants activated by p65/RelA concentration (i.e., inhibition) is presented as the ratio of the relative luciferase activity in the presence of DSIF RNAi to the activity in the presence of the parental vector pSuper. (B) Effect of DSIF knockdown by two distinct RNAis on the wild-type A20 promoter and a mutant (A20-TATA) which was converted into a canonical TATA box promoter by substitution of two nucleotides (bold letters) at −30 and −26 relative to the transcription start site, in the presence or absence of p65/RelA (suboptimal concentration). DSIF RNAi 1 is the one used for Fig. 1, and DSIF RNAi 2 is directed against a different region of the p160 subunit (see Materials and Methods). The inhibition results are presented as in panel A and are the averages of seven (DSIF RNAi 1) or six (DSIF RNAi 2) independent duplicate transfection experiments. Representative immunoblots showing down regulation of DSIF p160 by the two RNAis are shown on the right. (C) Responsiveness of the A20 mutant reporter genes to p65/RelA NF-κB protein.
FIG. 3.
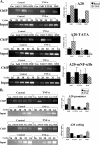
The core promoter controls differential occupancy of DSIF on A20 gene upon NF-κB induction. (A) The wild-type and mutant A20 promoters-driven reporter plasmids, as indicated, were each transfected into 293T cells. Cells were treated 24 h later with TNF-α for 1 h and then subjected to ChIP with the indicated antibodies and an irrelevant antibody as a control, followed by PCR analysis after normalization to the input (see Materials and Methods). The forward primer is specific to each of the promoters, and the reverse primer is derived from the proximal region of the luciferase gene. Representative ChIP results are shown on the left. Input DNA (0.1%) was subjected to an increasing number of PCR cycles in order to find the linear range to serve as a reference for the immunoprecipitation. Quantified results, normalized to the input, derived from two independent experiments are shown on the right. (B) Jurkat T cells were untreated or treated with TNF-α for 1 h, followed by ChIP assay with the indicated antibodies and an irrelevant antibody. The immunoprecipitated DNAs were subjected to PCR with primers specific to the promoter and beginning of each of the A20, β-actin, and BLR1 endogenous genes. Quantified results, normalized to the input, derived from three independent experiments are shown on the right.
FIG. 3.
The core promoter controls differential occupancy of DSIF on A20 gene upon NF-κB induction. (A) The wild-type and mutant A20 promoters-driven reporter plasmids, as indicated, were each transfected into 293T cells. Cells were treated 24 h later with TNF-α for 1 h and then subjected to ChIP with the indicated antibodies and an irrelevant antibody as a control, followed by PCR analysis after normalization to the input (see Materials and Methods). The forward primer is specific to each of the promoters, and the reverse primer is derived from the proximal region of the luciferase gene. Representative ChIP results are shown on the left. Input DNA (0.1%) was subjected to an increasing number of PCR cycles in order to find the linear range to serve as a reference for the immunoprecipitation. Quantified results, normalized to the input, derived from two independent experiments are shown on the right. (B) Jurkat T cells were untreated or treated with TNF-α for 1 h, followed by ChIP assay with the indicated antibodies and an irrelevant antibody. The immunoprecipitated DNAs were subjected to PCR with primers specific to the promoter and beginning of each of the A20, β-actin, and BLR1 endogenous genes. Quantified results, normalized to the input, derived from three independent experiments are shown on the right.
FIG. 4.
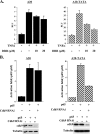
P-TEFb occupancy is the inverse of DSIF upon NF-κB induction. (A) ChIP of transfected A20 wild-type and mutant reporter genes, as described for Fig. 3A, using antibodies against Pol II, the cdk9 subunit of P-TEFb, and the p160 subunit of DSIF and an irrelevant antibody as a control. Representative results of ChIP and input are shown on the left, and quantified results, normalized to the input, derived from two independent experiments are shown on the right. (B) ChIP assay of the endogenous A20 gene from control Jurkat T cells or cells treated for 1 h with TNF-α as described for Fig. 3B, using antibodies to unphosphorylated Pol II, the cdk9 subunit of P-TEFb, serine 2-phosphorylated Pol II (Ser2), and the p160 subunit of DSIF and an irrelevant antibody as a control (Con). The immunoprecipitated DNAs were subjected to PCR with primers specific to the promoters or to internal regions of these genes (coding). Representative results of ChIP and input are shown on the left, and quantified results, normalized to the input, derived from two independent experiments are shown on the right.
FIG. 5.
Differential regulation of NF-κB by P-TEFb is core promoter type dependent. (A) 293T cells were transfected with the indicated reporters and 24 h later were treated either with TNF-α or with TNF-α plus increasing amounts of DRB for 4 h. The luciferase activity was normalized to the values of the cotransfected RSV promoter-driven Renilla reporter luciferase plasmid. The graphs show means and standard deviations of three duplicate experiments. (B) 293T cells were transfected with either wild-type A20 or A20-TATA mutant promoters together with p65/RelA expression vector and with either pSuper (parental vector) or cdk9 RNAi as indicated. Cells were harvested 48 h posttransfection, and luciferase activity was measured. Data are means and standard deviations of six independent duplicated experiments (left). A representative immunoblot showing cdk9 depletion in transfected cells is shown on the bottom.
FIG. 6.
ChIP analysis of other NF-κB target genes. The TATA-less IκBα and the TATA-containing RANTES genes were subjected to ChIP assay before or 1 h after TNF-α treatment as described for Fig. 3B. Antibodies to unphosphorylated Pol II, serine 2-phosphorylated Pol II (Ser2), and the p160 subunit of DSIF and an irrelevant control antibody (Con) were used for immunoprecipitation. The precipitated DNAs were subjected to PCR with primers specific to the promoters or the coding regions of the genes. Representative results of ChIP and input are shown on the left. Quantified results, normalized to the input, derived from two independent experiments are shown on the right.
FIG. 7.
Serine 2 phosphorylation of Pol II on the TATA promoter releases DSIF. Jurkat cells were untreated or treated with TNF-α without or with DRB (20 μM) for 1 h and then subjected to ChIP assay with antibodies to serine 2-phosphorylated Pol II (Ser2) and the p160 subunit of DSIF and an irrelevant antibody as a control (Con) followed by PCR analysis of the TATA-less IκBα and the TATA-containing RANTES promoters. Representative results from two experiments are shown. Quantified results, normalized to the input, derived from two independent experiments are shown on the right.
FIG. 8.
Dependency of the different NF-κB-responsive promoters on TAF1. (A) Wild-type A20 and A20-TATA mutant promoters were each transfected into 293T cells and analyzed by ChIP as described for Fig. 3A, using antibodies against TAF1 and Pol II and an irrelevant antibody as a control (Con). Representative results of ChIP and input are shown on the left, and quantified results, normalized to the input, derived from two independent experiments are shown on the right. (B) Fixed chromatin extract was prepared from Jurkat T cells induced by TNF-α for 1 h. ChIP assays were performed as described for Fig. 3B using antibodies to Pol II and TAF1 and an irrelevant antibody as a control. Representative results are shown on the left, and quantified results, normalized to the input (as in Fig. 4B) derived from two independent experiments on the right. (C) Temperature-sensitive hamster ts13 cells were cotransfected with the indicated reporter plasmids together with the p65/RelA expression plasmid. The cells were incubated at the permissive temperature (32°C) for 6 h, washed, and then separated into two groups. One was grown at 32°C and the second at the nonpermissive temperature (39.5°C) for an additional 48 h, after which the cells were harvested and luciferase activity measured. RSV promoter-driven Renilla reporter luciferase plasmids served to normalize transfection efficiency within each group, and c-_fos_-luciferase (TAF-independent promoter) served to normalize transfection efficiency between the groups. Results are means of four independent experiments, each with independent duplicates. (D) Hamster ts13 cells were grown at 32°C and then shifted to the nonpermissive temperature (39.5°C) for 6 h, followed by a 1-h treatment with mouse TNF-α. Total RNA was extracted and subjected to a quantitative RT-PCR assay to measure the mRNAs of A20, IκBα, and β-actin genes, using a Light Cycler system. Data are means and standard deviations of two independent experiments.
FIG. 8.
Dependency of the different NF-κB-responsive promoters on TAF1. (A) Wild-type A20 and A20-TATA mutant promoters were each transfected into 293T cells and analyzed by ChIP as described for Fig. 3A, using antibodies against TAF1 and Pol II and an irrelevant antibody as a control (Con). Representative results of ChIP and input are shown on the left, and quantified results, normalized to the input, derived from two independent experiments are shown on the right. (B) Fixed chromatin extract was prepared from Jurkat T cells induced by TNF-α for 1 h. ChIP assays were performed as described for Fig. 3B using antibodies to Pol II and TAF1 and an irrelevant antibody as a control. Representative results are shown on the left, and quantified results, normalized to the input (as in Fig. 4B) derived from two independent experiments on the right. (C) Temperature-sensitive hamster ts13 cells were cotransfected with the indicated reporter plasmids together with the p65/RelA expression plasmid. The cells were incubated at the permissive temperature (32°C) for 6 h, washed, and then separated into two groups. One was grown at 32°C and the second at the nonpermissive temperature (39.5°C) for an additional 48 h, after which the cells were harvested and luciferase activity measured. RSV promoter-driven Renilla reporter luciferase plasmids served to normalize transfection efficiency within each group, and c-_fos_-luciferase (TAF-independent promoter) served to normalize transfection efficiency between the groups. Results are means of four independent experiments, each with independent duplicates. (D) Hamster ts13 cells were grown at 32°C and then shifted to the nonpermissive temperature (39.5°C) for 6 h, followed by a 1-h treatment with mouse TNF-α. Total RNA was extracted and subjected to a quantitative RT-PCR assay to measure the mRNAs of A20, IκBα, and β-actin genes, using a Light Cycler system. Data are means and standard deviations of two independent experiments.
Similar articles
- The transcription elongation factors NELF, DSIF and P-TEFb control constitutive transcription in a gene-specific manner.
Fujita T, Piuz I, Schlegel W. Fujita T, et al. FEBS Lett. 2009 Sep 3;583(17):2893-8. doi: 10.1016/j.febslet.2009.07.050. Epub 2009 Aug 3. FEBS Lett. 2009. PMID: 19654008 - Positive transcription elongation factor b (P-TEFb) contributes to dengue virus-stimulated induction of interleukin-8 (IL-8).
Li LL, Hu ST, Wang SH, Lee HH, Wang YT, Ping YH. Li LL, et al. Cell Microbiol. 2010 Nov;12(11):1589-603. doi: 10.1111/j.1462-5822.2010.01493.x. Cell Microbiol. 2010. PMID: 20618343 - Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro.
Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Wada T, et al. EMBO J. 1998 Dec 15;17(24):7395-403. doi: 10.1093/emboj/17.24.7395. EMBO J. 1998. PMID: 9857195 Free PMC article. - Controlling the elongation phase of transcription with P-TEFb.
Peterlin BM, Price DH. Peterlin BM, et al. Mol Cell. 2006 Aug 4;23(3):297-305. doi: 10.1016/j.molcel.2006.06.014. Mol Cell. 2006. PMID: 16885020 Review. - Advances in NF-kappaB signaling transduction and transcription.
Xiao W. Xiao W. Cell Mol Immunol. 2004 Dec;1(6):425-35. Cell Mol Immunol. 2004. PMID: 16293211 Review.
Cited by
- Wide-scale analysis of human functional transcription factor binding reveals a strong bias towards the transcription start site.
Tabach Y, Brosh R, Buganim Y, Reiner A, Zuk O, Yitzhaky A, Koudritsky M, Rotter V, Domany E. Tabach Y, et al. PLoS One. 2007 Aug 29;2(8):e807. doi: 10.1371/journal.pone.0000807. PLoS One. 2007. PMID: 17726537 Free PMC article. - The Long Non-coding RNAs: Paramount Regulators of the NLRP3 Inflammasome.
Menon MP, Hua KF. Menon MP, et al. Front Immunol. 2020 Sep 25;11:569524. doi: 10.3389/fimmu.2020.569524. eCollection 2020. Front Immunol. 2020. PMID: 33101288 Free PMC article. Review. - RNA polymerase II pausing during development.
Gaertner B, Zeitlinger J. Gaertner B, et al. Development. 2014 Mar;141(6):1179-83. doi: 10.1242/dev.088492. Development. 2014. PMID: 24595285 Free PMC article. Review. - The elongation factor Spt5 facilitates transcription initiation for rapid induction of inflammatory-response genes.
Diamant G, Bahat A, Dikstein R. Diamant G, et al. Nat Commun. 2016 May 16;7:11547. doi: 10.1038/ncomms11547. Nat Commun. 2016. PMID: 27180651 Free PMC article. - Ready, pause, go: regulation of RNA polymerase II pausing and release by cellular signaling pathways.
Liu X, Kraus WL, Bai X. Liu X, et al. Trends Biochem Sci. 2015 Sep;40(9):516-25. doi: 10.1016/j.tibs.2015.07.003. Epub 2015 Aug 4. Trends Biochem Sci. 2015. PMID: 26254229 Free PMC article. Review.
References
- Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. - PubMed
- Basehoar, A. D., S. J. Zanton, and B. F. Pugh. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116:699-709. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources